Keywords
Pseudomonas aeruginosa; Multidrug-resistant; Serogroups; rpoB gene
Introduction
Antibiotics are usually used for the prevention and treatment of animal diseases in most of the livestock production systems to improve the efficiency of animal production [1]. These drugs are quickly excreted from the animal, others are not readily metabolized or excreted and so, their residues will persist in the animal tissues and hence enter the human food chain constituting health risks to the consumers [1,2]. At the level of important consumer animal products, residues of antibiotic can contribute to selection of resistant zoonotic bacteria (Salmonella, Campylobacter, E. coli), pathogens in humans as well as resistant commensal bacteria [1,3,4].
All these resistant commensal bacteria then constitute a reservoir of resistance genes that can be transferred to zoonotic bacteria [3]. Moreover, when these resistant digestive bacteria of animal origin are transmitted to humans, they can also transfer their resistance genes to bacteria in the commensal flora of human digestive tract [5].
Among these resistant bacteria, P. aeruginosa expresses a multiresistance to antibiotics and this resistance can be acquired (plasmids, transposons) or natural [6,7]. This resistance generally favors the involvement of P. aeruginosa in nosocomial infections, food poisoning and biofilms formation [8-10]. The formation of biofilms gives P. aeruginosa a high power of colonization, alteration of foodstuffs, and resistance to antiseptics, disinfectants and antibiotics [9,11,12].
The consequence is that P. aeruginosa is feared and is of concern both in the field of public health and in the agro-food industry [13]. Foods can play an important role in the propagation and antibiotic resistance of P. Pseudomonas strains [13]. Pseudomonas aeruginosa is also the cause of infection in some animals such as sheep, horses, bovines and dogs [6]. In humans, P. aeruginosa is the cause of dermatitis, meningitis, skin infections in severe burns, sepsis, and nosocomial infections of the urinary tract [8,14,15]. This bacterium is also responsible for ophthalmological and digestive infections [15]. The prevalence of P. aeruginosa infections is 11.5% in Europe and 17% in developing countries.
Despite the different therapeutic advances, the mortality of P. aeruginosa infections remains high, oscillating at around 30% [16]. Also, 20% of nosocomial infections are due to P. aeruginosa. In patients receiving mechanical ventilation, 89% are colonized by P. aeruginosa, 17% of which develop pneumonia due to natural or acquired resistance [6,12]. The emergence of this bacteria and its spread in human populations is a public health problem and results in increasing levels of resistance to antibiotics [17].
In addition to a rather impressive arsenal of virulence factors, P. aeruginosa has mechanisms to withstand all currently available antibiotics [18,19]. In Pseudomonas aeruginosa, this resistance is most often related to chromosomal mechanisms (alteration of the porcine OprD) or to the production of enzymes hydrolyzing carbapenems [6,12]. Since their first use, beta-lactams have been the most prescribed and widely used antibiotic family in the world due to their broad spectrum of action, low toxicity, efficacy and low cost. In Africa, many cases of multi-resistant bacteria are reported [11,20,21].
In Côte d'Ivoire, although some studies have shown the characterization of Pseudomonas aeruginosa associated with nosocomial infections [17,22] there is no data on the resistance potential of strains of Pseudomonas aeruginosa contaminating animal products. The presence of antibiotics in animal product is associated with several adverse public health effects including hypersensitivity, tissue damage, gastrointestinal disturbance and bacterial resistant strain. Protection of public health against possible harmful effects of antibiotic residues is a very important problem.
This study aimed to evaluate the multidrug-resistance of Pseudomonas aeruginosa isolated from bovine meat, smoked fish and fresh fish.
Materials and Methods
Study area
Sampling of animal products was carried out from February 2015 to November 2015 in five (5) communes of Abidjan, including Port-Bouet, Abobo, Yopougon, Adjamé and Binger-ville. Fresh and smoked fish samples were taken only from sales markets. In addition to these sales sites, bovine meat was also harvested at the slaughterhouse.
Sampling
A total of five hundred (500) samples about 200 grams consisted of 230 bovine meat, 130 fresh fish, and 140 smoked fish were taken. Animals samples were carried to the laboratory of the Department of Bacteriology and Virology of Institute Pasteur of Côte d'Ivoire in pre-identified Stomacher bags, stored at 4°C, and analyzed within 30 minutes of collection.
Isolation and identification of Pseudomonas aeruginosa
Twenty five g of the flesh of each bovine meat, fresh fish and smoked fish sample were homogenized in 225 ml peptone water, and then, serial decimal dilutions were prepared. Amount of 0.1 ml of each dilution was spread on the selective medium Pseudomonas Cetrimide agar (PCA) using a spreading technique. Plates were incubated at 44?C for 18-24 hours and observed for suspected colonies of P. aeruginosa. The isolates were identified as P. aeruginosa in the Bacteriology and Virology laboratory using the API 20NE (bioMérieux, Marcy l’Etoile, France) and the API database. Molecular markers based on the analysis of rpoB household genes were used to characterize and confirm the identity of presumptive P. aeruginosa isolates.
Susceptibility testing
Antimicrobial susceptibility was determined by diffusion method on Mueller-Hinton agar (MHA; Bio-Rad, Marnes-La-Coquette, France), and the data were interpreted according to the CA-SFM/ EUCAST recommendations [23]. The antibiotics tested and their sensidisk concentrations were ticarcillin (TIC; 75 μg), ticarcillinclavulanic acid (TCC; 75-10 μg), aztreonam (ATM; 30 μg), cefepime (FEP; 30 μg), ceftazidine (CAZ; 10 μg), ciprofloxacin (CIP; 100 μg), colistin (CST; 10 μg), imipenem (IPM; 10 μg), piperacillin (PIP; 100 μg), fosfomycin (FOS; 200 μg) and kanamycin (K; 30 μg).
Bacterial suspension was prepared by touching the top of the isolated colonies of test organism grown on nutrient agar with sterile wire loop and suspending in a tube containing 2-3 ml of 0.85% sterile saline, turbidity was adjusted to 0.5 McFarland standards. Suspension was evenly spread on Mueller Hinton agar (MHA; Bio-Rad, Marnes-La-Coquette, France) by using a sterile cotton swab. Antibiotics discs were placed on the plate with the help of sterile forceps. After the disks were placed, the plate were inverted and incubated at 37°C for 16 to 18 hours. Zones of inhibition were measured after overnight incubation according to the CLSI guideline. All strains showing resistance or intermediate resistance were assumed resistant. Pseudomonas aeruginosa ATCC 27853 was used as a control strain for the verification of our procedures.
Definition of multidrug-resistant (MDR) isolates
Multidrug-resistant (MDR) P. aeruginosa isolates were defined as resistant to at least three of the following antibiotics: ceftazidim, imipenem, cefepime, ticarcillin and ciprofloxacin [24], using the conventional serial agar dilution method. The minimal inhibitory concentrations were interpreted according to the CA-SFM / EUCAST [23].
Genomic DNA extraction
A single bacterial colony was inoculated into 5 ml (LB) and grown for 18 h at 30°C. Saturated culture was harvested with centrifugation for 3 min at 14,000 rpm. The cell pellet was suspended and lysed in 200 μl of lysis buffer (40 mM Tris-acetate pH 7.8, 20 mM sodium-acetate, 1 mM EDTA, 1% SDS) by vigorous pipetting. To remove most proteins and cell debris, 66 μl of 5M NaCl solution was added and mixed well, and then the viscous mixture was centrifuged for 10 min at 14,000 rpm at +4°C. An equal volume of chloroform was added to the clear supernatant. Following centrifugation at 14,000 rpm for 3 min, the extract supernatant was precipitated with 100% EtOH, washed twice with 70% EtOH, dried and redissolved in 50 μl 1x TE buffer [25].
PCR amplification. Single PCR for characterization of rpoB gene was carried out with a total volume of 25μl consisted of 16 μl of sterile Milli-Q water (milli-Q™, Millipore Corporation, USA), 5μl of 5XTP, 1.5 μl of MgCl2 (2 mM), 0.2 μl of dNTPs (10 mM), 0.1μl of each primer (20 mM) (Integral DNA Technology, California, USA) (rpoB F: Order No. 2512433, Ref. No. 70393602; rpoB R : Order No. 2512433, Ref. No. 70393603), 0.1 μl of Go tag polymerase (Promega Corporation, Madison, WI 53711-5399, USA) and 2 μl of DNA matrix. DNA rpoB region amplification was performed using the primer set rpoB F’ (5’-CAGTTCATGGACCAGAACAACCCG-3’) and rpoB R’ (5’ACGCTGGTTGATGCAGGTGTTC-3’), aligning on positions 1552 and 2298 of the rpoB gene sequence of Pseudomonas aeruginosa UCBPP-PA14 (CP000438).
The rpoB DNA was amplified using the following protocol: initial denaturation at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 1min, annealing at 58°C for 1 min and extension at 72°C for 2 min, with a single final extension of 7 min at 72°C. Amplification was performed with 10 μl of PCR products which were separated in 1.5% agarose gel for 30 min at 120V. After amplification, the agarose gel was put in ethidium bromide (0.5 μg/ml) and detected by Molecular Imager Gel DocTM EZ (Bio- Rad, USA).
Serotyping of Pseudomonas aeruginosa isolates
The O-serotypes were determined by a slide agglutination test using four pools (OMA, OMC, OME, and OMF) and 20 monovalent antisera, O1 to O20 (Sanofi Diagnostics Pasteur), according to the manufacturer’s recommendations.
Statistical analysis
Data analysis was performed using the statistical package for the Social Sciences (SPSS), version 20.0 (IBM SPSS, Chicago, IL, United States of America). The chi-squared test was used at 5% significance level.
Results
Pseudomonas aeruginosa strains detected by API 20NE and rpoB
On a set of 225 presumptive isolates of Pseudomonas, 205 (91.1%) belonging to Pseudomonas aeruginosa were identified by API 20NE (Figure 1 and Table 1). A total of 204 (99.5%) strains were confirmed by the rpoB gene as belonging to Pseudomonas aeruginosa out of the 205 (91.1%) isolates previously identified by API 20NE as belonging to Pseudomonas aeruginosa.
| Identification |
Number of isolatesPresumptiveP. aeruginosa N=225 |
| |
Confirmedspecies |
Effective |
Percentage |
| Biochemical |
P. aeruginosa |
205 |
91.1 |
| using API 20NE |
|
|
| MolecularusingrpoB |
P. aeruginosa |
204 |
99.5 |
Table 1: Frequency of strains confirmed by the API 20NE and rpoB genes
Figure 1: RpoB profiles of Pseudomonas aeruginosa isolates. Lanes 1-5; 7-16: Presence of Pseudomonas aeruginosa in analyzed samples; Lanes 6: Absence of Pseudomonas aeruginosa in analyzed samples C+: Positive control, Pseudomonas aeruginosa ATCC 27853; C-: Negative control; M: Marker Gene Ruler 250 bp (Bench Top, 1kb DNA Ladder, Promega Corporation, USA).
Prevalence of multidrug-resistant (MDR) Pseudomonas aeruginosa isolates
The results showed that 181 strains of Pseudomonas aeruginosa or 88.7% of the strains isolated with a total prevalence of 36.2% were multidrug-resistant (Table 2). The prevalence of Pseudomonas aeruginosa multidrug-resistant (PAMDR) was high in beef (47.8%) followed by fresh fish (33.1%) and lower in smoked fish (20.0%) (Table 2).
| Animal products |
Effective of sample |
Strains of P. aeruginosaisolated |
Strains of P. aeruginosa MDR |
Frequency of P. aeruginosa MDR % |
Prevalence of P. aeruginosa MDR % |
| Bovine meat |
230 |
122 |
110 |
53.9 |
47.8 |
| Freshfish |
130 |
49 |
43 |
21.1 |
33.1 |
| Smokedfish |
140 |
33 |
28 |
13.7 |
20 |
| Total |
500 |
204 |
181 |
88.7 |
36.2 |
Table 2: Prevalence of Pseudomonas aeruginosa multidrug-resistant MDR.MDR: Multidrug-resistant.
Frequency of multidrug resistant isolates
The percentage of resistance showed by P. aeruginosa isolates were 98.4% for aztreonam, 51.4% ticarcillin + clavulanic acid, 50.4% ticarcillin, 31.4% piperacillin, 33.6% ciprofloxacin, 17.0% cefepime, 6.9% ceftazidime, 7.2% imipenem, 4.5% colistin and 0.0% fosfomycin (Figure 2).
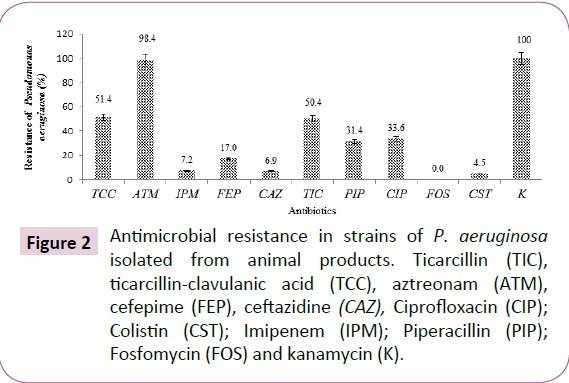
Figure 2: Antimicrobial resistance in strains of P. aeruginosa isolated from animal products. Ticarcillin (TIC), ticarcillin-clavulanic acid (TCC), aztreonam (ATM), cefepime (FEP), ceftazidine (CAZ), Ciprofloxacin (CIP); Colistin (CST); Imipenem (IPM); Piperacillin (PIP); Fosfomycin (FOS) and kanamycin (K).
Resistance profile of P. aeruginosa according to the original animal product
All strains of Pseudomonas aeruginosa isolated from bovine meat, fresh and smoked fish expressed resistance to almost all antibiotics. The strains of Pseudomonas aeruginosa isolated from smoked fish are the most resistant to ticarcillin/ticarcillin + clavulanic acid with 68.4% (Table 3). Resistance to these two antibiotics was 52.5% and 50.0% respectively for strains isolated from fresh fish and 30.2% and 35.7% respectively for those isolated from bovine meat. The same strains isolated from smoked fish were also more resistant to cefepime with 23.7%, while those isolated from bovine meat and fresh fish were less resistant to this antibiotic with 17.5% and 10%, respectively. The resistance to ceftazidime and imipenem of strains isolated from bovine meat was 7.9% and 8.7% respectively (Table 3).
| Type of Sample |
Percentage of strainsresistance to antibiotics % |
| |
TCC |
ATM |
IPM |
FEP |
CAZ |
TIC |
FOS |
PIP |
CIP |
CST |
K |
| Bovine meat |
35.7 |
97.6 |
8.7 |
17.5 |
7.9 |
30.2 |
0 |
34.2 |
35.4 |
5.1 |
100 |
| Freshfish |
50 |
97.5 |
7.5 |
10 |
7.5 |
52.5 |
0 |
32 |
33.2 |
5.3 |
100 |
| Smokedfish |
68.4 |
100 |
5.3 |
23.7 |
5.3 |
68.4 |
0 |
28 |
32.2 |
3.2 |
100 |
| Resistanceaverage |
51.4±23.1 |
98.4±1.6 |
7.2±2.4 |
17±0.7 |
6.9±1.8 |
50.4±27.0 |
0± 0 |
31.4±4.3 |
33.6±2.2 |
4.5±1.3 |
100±0 |
| TIC:Ticarcillin;TCC: Ticarcillin-Clavulanic;ATM:AcidAztreonam;FEP: Cefepime;CAZ: Ceftazidine;CIP: Ciprofloxacin;CST: Colistin;IPM: Imipenem;PIP: Piperacillin; FOS:Fosfomycin; and K: Kanamycin K. |
Table 3: Resistance of P. aeruginosa according to animal product.
This resistance to ceftazidime/imipenem was less important for strains isolated respectively from fresh fish with 7.5% and smoked with 5.3% for each antibiotic (Table 3). The resistance of Pseudomonas aeruginosa strains isolated from beef, fresh and smoked fish to colistin ranges from 3.2% to 5.3%. Resistance to the three types of products varied from 35.4% to 32.2% for ciprofloxacin and from 34.2% to 28% for piperacillin. All strains of Pseudomonas aeruginosa isolated from animal products were susceptible to fosfomycin. However, they showed more than 97% resistance to aztreonam (Table 3).
Phenotype of multidrug resistant isolates
A total of seven (7) antibiotic resistance profiles (phenotypes) were identified in the isolates (Table 4). The majority were phenotype II, I, and phenotype III with respectively 36.7%, 23.9% and 12.3% resistance to antibiotics. The other phenotypes exhibited resistance frequencies of less than 10%.
| Phenotype |
Number of Pseudomonas aeruginosastrains N |
Frequency of resistantisolates |
Resistance to Antibiotics |
| I |
47 |
23.9 |
TIC TCC FEP CAZ IPM ATM CIP PIP K FOS CST |
| II |
72 |
36.7 |
TIC TCC FEP CAZ IPM ATM CIP PIP |
| III |
24 |
12.3 |
TIC TCC FEP CAZ IPM PIP |
| IV |
16 |
8.2 |
TIC FEP CAZ IPM CIP |
| V |
12 |
6.1 |
TCC CAZ IPM CIP |
| VI |
10 |
5.1 |
FEP CAZ |
| VII |
15 |
7.7 |
IPM |
| Total |
196 |
100 |
|
| Ticarcillin TIC, ticarcillin-clavulanicacid TCC, aztreonam ATM, cefepime FEP, Ceftazidine CAZ, Ciprofloxacin CIP, Colistin CST, Imipenem IPM, Piperacillin PIP, Fosfomycin FOS and Kanamycin K. |
Table 4: Pseudomonas aeruginosaantibiotic resistance phenotype isolated.
Pseudomonas aeruginosa resistant to imipenem
A total of fifteen (15) strains of P. aeruginosa isolated from animal products were resistant to imipenem. Of the fifteen (15) strains of P. aeruginosa, nine (9) or 60% of the isolated strains of bovine meat showed an average inhibitory concentration greater than 32 mg/L (Table 5, Figure 3a and 3b). The mean minimum inhibitory concentrations of imipenem for strains isolated from fresh fish 4 (26.7%) and smoked 2 (13.3%) were respectively 32 mg/L and 24 mg/L (Table 5).
| Type of Sample |
Bovine meat |
Fresh fish |
Smoked fish |
Total |
| Number N of Pseudomonas aeruginosa strains resistance to imipenem IPM |
9 |
4 |
2 |
15 |
| Percentage of Pseudomonas aeruginosa strain resistance to imipenem |
60.00% |
26.70% |
13.30% |
100% |
| MIC average of imipenem in mg/L |
≥32 |
32 |
24 |
|
| MIC: MinimalesInhibitrices Concentrations; imipenem IPM |
|
|
|
|
Table 5: Minimum inhibitory concentration of imipenem for P. aeruginosa strains.
Figure 3a: Multi-resistance to antibiotics of Pseudomonas aeruginosa isolated from bovine meat.
Figure 3b: Resistance to imipenem of Pseudomonas aeruginosa isolated from bovine meat with MIC ≥ 32 mg/L.
Resistance and serogroups of Pseudomonas aeruginosa
The results we reported that Pseudomonas aeruginosa serogroups O1, O2, O4 and O10 were sensitive to the majority of the tested antibiotics (ticarcillin, ticarcillin-clavulanic acid, cefepime, ceftazidime, ciprofloxacin, imipenem, piperacillin and fosfomycin) (Table 6).
| ATB % |
Resistance and serogroups of Pseudomonas aeruginosaisolated N=204 |
| |
NS |
O1 |
O10 |
O11 |
O12 |
O15 |
O16 |
|
O2 |
O4 |
O5 |
O7 |
O8 |
O9 |
| TCC |
62 |
0 |
0 |
60 |
50 |
20 |
27 |
|
0 |
0 |
33 |
66 |
40 |
33 |
| ATM |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
|
100 |
100 |
100 |
100 |
100 |
93 |
| IPM |
10 |
0 |
0 |
20 |
0 |
0 |
9 |
0 |
|
0 |
33 |
15 |
0 |
8 |
| FEP |
25 |
0 |
0 |
20 |
0 |
0 |
27 |
0 |
|
0 |
33 |
15 |
0 |
12 |
| CAZ |
12 |
0 |
0 |
20 |
0 |
0 |
9 |
0 |
|
0 |
22 |
15 |
0 |
13 |
| TIC |
37 |
0 |
0 |
40 |
50 |
25 |
36 |
|
0 |
0 |
33 |
55 |
20 |
20 |
| PIP |
20 |
0 |
0 |
45 |
30 |
20 |
25 |
|
0 |
0 |
35 |
40 |
30 |
26 |
| CIP |
18 |
0 |
0 |
49 |
30 |
18 |
28 |
|
0 |
0 |
32 |
42 |
32 |
24 |
| FOS |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
0 |
0 |
0 |
0 |
0 |
0 |
| CST |
6 |
8 |
10 |
8 |
10 |
8 |
8 |
|
10 |
12 |
12 |
10 |
6 |
8 |
| K |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
|
100 |
100 |
100 |
100 |
100 |
100 |
Table 6: Distribution of Pseudomonas aeruginosa serogroups according to the antibiotics.
In addition, strains belonging to serogroups O1, O2, O4, O8, O10, O12 and O15 were both susceptible to cefepime, ceftazidim and imipenem (Table 6). However, strains of serogroups O5, O7, O9, O11, O16 and non-serotyping strains (NS) were resistant to the majority of antibiotics tested (Table 6). Pseudomonas aeruginosa strains mainly resistant to ticarcillin, ticarcillin-clavulanic acid, cefepime, imipenem, ceftazidime, ciprofloxacin and piperacillin were O5, O7 and O11 serogroups (Table 6 and Figure 4).
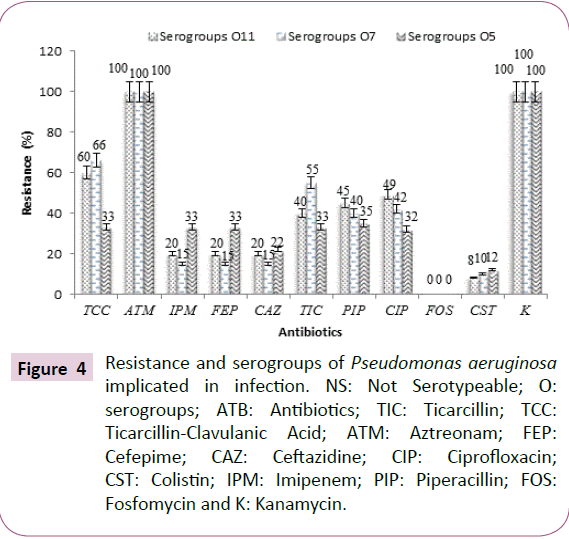
Figure 4: Resistance and serogroups of Pseudomonas aeruginosa implicated in infection. NS: Not Serotypeable; O: serogroups; ATB: Antibiotics; TIC: Ticarcillin; TCC: Ticarcillin-Clavulanic Acid; ATM: Aztreonam; FEP: Cefepime; CAZ: Ceftazidine; CIP: Ciprofloxacin; CST: Colistin; IPM: Imipenem; PIP: Piperacillin; FOS: Fosfomycin and K: Kanamycin.
Discussion
The high molecular identification rate showed that genomic studies are needed to confirm the exact taxonomic position of Pseudomonas aeruginosa. The results of this study confirmed that a correct identification and characterization of Pseudomonas aeruginosa can only be achieved by combining cultural, biochemical and molecular tests.
Analysis of the results showed also that 181 strains of Pseudomonas aeruginosa or 88.7% of the strains isolated with a total prevalence of 36.2% were multi resistant to antibiotics. These results are in agreement with those obtained by Mohammad et al. [26]. In fact, they isolated P. aeruginosa multi-drug resistant from different infection sites. This high multi resistance could be due to the production of hydrolytic enzymes and the acquisition of resistance mechanisms by Pseudomonas aeruginosa strains [6,12,15]. Multidrug-resistance of these Pseudomonas aeruginosa strains could also be due to residues of antibiotics used in an uncontrolled manner in the production of these animal products [1,27]. The prevalence may be due to an increase in antibiotic treatments. This prevalence of Pseudomonas aeruginosa multidrug-resistant (PAMR) was raised for beef (47.8%) followed by fresh fish (33.1%) and lowest for smoked fish (20.0%).
This high prevalence of multidrug-resistant Pseudomonas aeruginosa in fresh produce could be explained by the fact that Pseudomonas aeruginosa colonizes more moist sites [28,29]. The low prevalence of multidrug-resistant Pseudomonas aeruginosa level smoked fish could also be explained by the fact that during fish smoking, the smoke particles absorbed by fish especially inhibit bacterial growth on the product surface [30].
The results also showed that the total antibiotic resistance of P. aeruginosa strains of animal origin was 98.4%, 51.4% and 50.4% respectively for aztreonam, ticarcillin-clavulanic acid + and ticarcillin. The high resistance of these strains to aztreonam and penicillin could be acquired (plasmids, transposons) and also could explain an increased impermeability of the outer membrane or in the production of inactivating enzymes according to Kumar and Schweizer [31].
The resistance of P. aeruginosa strains to cefepime (17.0%), ceftazidim (6.9%) and imipenem (7.2%) was higher than the resistance to colistin (4.5%) and fosfomycin (0%). This resistance of Pseudomonas aeruginosa to cephalosporin (ceftazidim and cefepime) may be due to chromosomal mechanisms (impaired OprD porin in Pseudomonas aeruginosa), the association of resistance mechanisms (Extended Spectrum Beta-Lactamase (ESBL) [6,12].
These resistances to cephalosporin were observed by Aman et al. [20] who found cefepime (34%) and ceftazidim (37%) higher resistance levels than in this study. This difference in resistance to cephalosporin could also be justified by the fact that Aman et al. [20] worked on environmental strains isolated from fresh water contaminated with domestic wastewater. The resistance to imipenem is related to the reduction of permeability by loss of porin D2 (OprD) according to Bricha et al. [6]. Thus, it is linked to an enzymatic mechanism by the production of carbapenemases which induce high-level resistance to all betalactamins [19,32]. This loss of porin is responsible for the increase in MIC ranging from 24 to 32 mg/L in the strains from animal origin, making the strains resistant to carbapenems according to Bricha et al. [6].
The results also indicate that all strains of Pseudomonas aeruginosa isolated from bovine, fresh and smoked fish expressed resistance to almost all antibiotics. The strains of Pseudomonas aeruginosa isolated from smoked fish are the most resistant to ticarcillin/ticarcillin + clavulanic acid with 68.4%. Resistance to these two antibiotics was 52.5% and 50.0% respectively for strains isolated from fresh fish and 30.2% and 35.7% respectively for those isolated from bovine meat.
The same isolated strains of smoked fish were also more resistant to cefepime with 23.7% compared to 17.5% and 10% respectively for beef and fresh fish. This resistance to cefepime in smoked fish may be due to a transfer of resistance gene from environmental strains to animal strains. Indeed, this type of fish is most often exposed for a long time in the environment which could favor the transfer of resistance gene from environmental strains to animal strains. The resistance to ceftazidim and imipenem of strains isolated from bovine meat was 7.9% and 8.7% respectively. This resistance to ceftazidim and imipenem was less important for strains isolated respectively from fresh and smoked fish.
This decrease in resistance to fresh and smoked fish strains could be explained by the fact that these two types of products were less in contact with environmental strains [31]. However, the resistance to ceftazidim observed in beef strains may be due to prior contamination of the animal by certain environmental strains prior to slaughter, during evisceration or at sales sites. The resistance of Pseudomonas aeruginosa strains isolated from beef, fresh and smoked fish to colistin ranges from 3% to 5.3%. These resistance levels of Pseudomonas aeruginosa strains obtained for colistin are contrary to the study carried out by Akhi et al. [33] who reported that all P. aeruginosa isolates were susceptible to colistin. This contradiction may be due to the fact that these authors have worked on clinical samples and not of animal origin [26].
It may also be explained by the sensitivity of methods, size and type of sample [34]. These strains of Pseudomonas aeruginosa isolated from animal products showed no resistance to fosfomycin. However, all resistance of P. aeruginosa to beta-lactams could be justified by the presence of the exopolysaccharide matrix which could slow the penetration of antibiotics and biocides [6,35]. These resistances to beta-lactams may be related to the presence of certain serotypes [35]. The strains of Pseudomonas aeruginosa, mainly resistant to ticarcillin, ticarcillin-clavulanic acid, cefepime, imipenem, ceftazidime, ciprofloxacin and piperacillin, were O5, O7 and O11 serogroups. These results show that serogroups O9, O16 and mainly serogroups O5, O7 and O11 may be associated with serious infections.
The results of this study indicate also the emergence of multidrug-resistant strains of Pseudomonas aeruginosa in the animal chain. Multidrug-resistant is a potential health threat to consumers. Studies have shown that multi-resistance of strains is an extremely important public health problem [4]. Therefore, bovine meat and fish must be kept clean and at low temperature to reduce or delay the growth of bacteria. Good hygienic practices and appropriate handling are necessary to prevent food poisoning associated with the consumption of bovine meat and fish [4,28]. There is an urgent need for a surveillance system for antibacterial drugs used in lifting and aquaculture practices to prevent multiresistance.
There is urgent need for a monitoring system of antibacterial drugs that are being used in lifting and aquaculture practices [4].
Conclusion
The study indicated the presence in various animal products, in particular bovine meat, fresh fish and smoked fish of multidrugresistant strains of Pseudomonas aeruginosa. She noted the level of resistance of these strains to the commonly prescribed and used antibiotics. The development of resistances, the incessant appearance of new mechanisms and the complexity of multiresistance phenotypes, require an improvement in the food chain management of the products analyzed to prevent the spread of multidrug-resistance or reduce the risk of infections. Bacterial contamination of fish and bovine meat is a cause of spoilage and is of particular importance if the fish and bovine meat is contaminated with pathogenic bacteria, which causes food poisoning or even death amongst consumers.
Acknowledgements
The authors thank the INRS-Institut Armand Frappier of Canada and the Pasteur Institute of Paris and the Institut Pasteur of Cote d'Ivoire for their advice and excellent technical assistance.
Declaration of Interest
The authors declare that there is no conflict of interest.
Funding Sources
There has been no source of funding.
19290
References
- Abdul SM, Muhammad K, Aijaz HS, Muhammad GS (2014) Antibiotic residues detection in raw beef meat sold for human consumption in sindh, Pakistan. International Journal of Research in Applied, Natural and Social Sciences 2: 15-20.
- Babapour A, Azami L, Fartashmehr J (2012) Overview of antibiotic residues in beef and mutton in Ardebil, North West of Iran. World Applied Science Journal 19: 1417-1422.
- Bibbal D (2008) Impact of beta-lactams on the emergence of resistant enterobacteria in the digestive flora in pigs: characterization and prevention strategy. University of Toulouse, Toulouse France 125.
- Tiamiyu AM, Soladoye MO, Adegboyega TT, Adetona MO (2015) Occurrence and Antibiotic Sensitivity of Bacterial Strains Isolated from Nile Tilapia, Oreochromis niloticus Obtained in Ibadan, Southwest Nigeria. Journal of Biosciences and Medicines 3: 19-26.
- Phillips IM, Casewell T, Cox B, De Groot C, Friis R, et al. (2004) Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J Antimicrob Chemother 53: 28-52.
- Bricha S, Ounine K, Oulkheir S, Haloui NEEL, Attarassi B (2009) Virulence factors and epidemiology related to Pseudomonas aeruginosa. Tunisian Journal of Infectious Diseases 2: 7-14.
- Sheetal S, Srivastava P (2016) Resistance of Antimicrobial in Pseudomonas aeruginosa. International Journal of Current Microbiology and Applied Sciences 5: 121-128.
- Mitov I, Tanya S, Boyka M (2010) Prevalence of virulence genes among bulgarian nosocomial and cystic fibrosis isolates of Pseudomonas aeruginosa. Brazilian Journal of Microbiology 41: 588-595.
- Bonkat G, Widmer AF, Rieken M, van der Merwe A, Braissant O, et al. (2013) Microbial biofilm formation and catheter associated bacteriuria in patients with suprapubic catheterisation. World Journal of Urology 31: 565-571.
- Tremblay YDN, Lamarche D, Chever P, Haine D, Messier S, et al.(2014) Characterization of the ability of coagulase-negative staphylococci isolated from the milk of Canadian farms to form biofilms. J Dairy Sciences 96: 234-246.
- Meseret M, Solomon A, Gebre K (2014) Antimicrobial Drug Resistance and Disinfectants Susceptibility of Pseudomonas aeruginosa Isolates from Clinical and Environmental Samples in Jimma University Specialized Hospital, Southwest Ethiopia. American Journal of Biomedical and Life Sciences2: 40-45.
- Syed H, Abidi KA, Fatin Z, Nouman M,Shahana UK (2016) Rabbit Serum Raised against P. aeruginosa Biofilm Exhibit Significant Biofilm Reduction and Removal Activity. International Journal of Current Microbiology Applied Sciences 5: 275-279.
- Virupakshaiah DBM, Hemalata VB (2016) Molecular identification of Pseudomonas aeruginosa from food borne isolates. International Journal of Current Microbiology Applied Sciences 5: 1026-1032.
- Maria JA, Joa CM, Barreira IC, Luis T, Liliana P, et al. (2014) Propensity for biofilm formation by clinical isolates from urinary tract infections: developing a multifactorial predictive model to improve antibiotherapy. Journal of Medical Microbiology63: 471-477.
- Rostamzadeh Z, Mohammadian M, Rostamzade A (2016) Investigation of Pseudomonas aeruginosa Resistance Pattern against Antibiotics in Clinical Samples from Iranian Educational Hospital. Advances in Microbiology 6: 190-194.
- Lisboa T, Waterer G, Rello J (2010) We should be measuring genomic bacterial load and virulence factors.Critical Care Medicine 38: 656-662.
- Guessennd NK, Ouattara MB, Ouattara ND, Nevry RK, Gbonon V, et al. (2013) Study of the multi-resistant bacteria of the hospital effluents of a hospital and university center (CHU) of the city of Abidjan (Côte d'Ivoire). Journal of Applied Biosciences 69: 5456-5464.
- Meenakumari S, Verma S, Absar A, Chaudhary A (2011) Antimicrobial susceptibility pattern of clinical isolates of Pseudomonas aeruginosa in an Indian cardiac hospital. International Journal of Eng Sci Technol 3: 7117-7124.
- Edit K, Sándor S, Gyula D, Júlia R, Balázs K, et al. (2016) Pathogenic and phylogenetic features of 2 multi-resistant Pseudomonas aeruginosa strains originated from remediated sites. International Journal of Occupational Medicine and Environmental Health 29: 503-516.
- Aman U, Rabia D, Ghadir A, Safia A (2012) Prevalence of antimicrobial resistant Pseudomonas aeruginosa in fresh water spring contaminated with domestic sewage. Journal of Biological and Food Science Research 1: 19-22.
- Hafsat AG, Yaqub AG, Abubakar S, Isa AG, Roy BB (2015) Multi-Drug Resistant Bacteria Isolated from Fish and Fish Handlers in Maiduguri, Nigeria. International Journal of Animal and Veterinary Advances 7: 49-54.
- Zinzendorf NY, Ouassa T, Agbessi BT, Kouassi KM, Ekra D, et al.(2009) Risk factors and microbial etiologies of nosocomial infections at the chu of Treichville, Abidjan (Côte d’Ivoire). Journal of science and pharmaceutical biology 10: 56-64.
- CA-SFM/EUCAST (2015) European Committee on Antimicrobial Susceptibility Testing and Antibiogram. Committee of the French Society of Microbiology 158.
- Lanotte PWS, Mereghetti L, Dartiguelongue N, Rastegar-Lari A, Goudeau A, et al. (2009) Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins. Journal of Medical Microbiol 53: 73-81.
- Chen WP, Kuo TT (1993) A simple and rapid method for preparation of gram-negative bacterial genomic DNA. Nucleic Acids Research 21: 22-60.
- Mohammad YM, Rahman P, Naser A, Reza G, Hossein BB (2016) Colistin, an option for treatment of multiple drug resistant Pseudomonas aeruginosa. Physiology and Pharmacology 20: 130-136.
- Farzanan AN, Nasrin AB, Atiqur RA, Habibur R, Firoz AM (2015) Occurrence of Antibiotic Resistant Bacteria in Pond Water Associated with Integrated Poultry-Fish Farming in Bangladesh. Sains Malaysiana 44: 371-377.
- Ibrahim KMD, Mosharraf HMD, Sharif NMD, Sherajul IMD, Kabir M (2015) Comparative efficiency of some commercial antibiotics against Pseudomonas infection in fish.International Journal of Fisheries and Aquatic Studies 2: 114-117.
- Al-Zaidi JR (2016) Antibiotic susceptibility patterns of Pseudomonas aeruginosa isolated from clinical and hospital environmental samples in Nasiriyah, Iraq. African Journal of Microbiology Research 10: 844-849.
- Idah PA, Nwankwo I (2013) Effects of smoke-drying temperatures and time on physical and nutritional quality parameters of Tilapia (Oreochromis niloticus). International Journal of Fisheries and Aquaculture 5: 29-34.
- Kumar A, Schweizer HP (2005) Bacterial resistance to antibiotics: active ef?ux and reduced uptake. Advanced Drug Delivery Research 57: 1486-1513.
- Ekrem K, Rokan DK (2014) Antibiotic susceptibility patterns of Pseudomonas aeruginosa strains isolated from various clinical specimens. Sky Journal of Microbiology Research 2: 13-17.
- Kumar A, Schweizer HP (2005) Bacterial resistance to antibiotics: active ef?ux and reduced uptake. Advanced Drug Delivery Research 57: 1486-1513.
- Akhi MT, Ghotaslou R, Beheshtirouy S, Asgharzadeh M, Pirzadeh T, et al. (2015) Antibiotic Susceptibility Pattern of Aerobic and Anaerobic Bacteria Isolated From Surgical Site Infection of Hospitalized Patients. Jundishapur J Microbiol 8: e20309.
- Shewatatek G, Gizachew T, Molalegne B, Terefe G (2014) Drug sensitivity of Pseudomonas aeruginosa from wound infections in Jimma University specialized hospital, Ethiopia. Journal of Medical Science Research 3: 13-18.
- Greta L,Gjyle MO,Rrezarta B,Arsim K, Elvir A, et al. (2016) Antimicrobial Resistance Profile And Serotyping of Pseudomonas aeruginosa in University Clinical Centre Of Kosovo. Acta Medica Mediterranea 32: 82-91.










