Introduction
Postoperative wound infections are infections that occur within 30 days of the operation or within 1 year of operation if an implant is left in place and if the infections appear to be related to the surgery [1]. The infection is associated with the intensity of bacterial contamination of the wound at surgery or later in wards during wound care that follows interference with the skin barrier [2]. The third most commonly reported nosocomial infection is postoperative wound infections which accounts for approximately a quarter of all nosocomial infections [3]. It has been responsible for the increasing cost, morbidity and mortality related to surgery and continues to be a major problem even in hospitals with modern facilities and standard protocols of preoperative safety measures and antibiotic prophylaxis [1,3].
Since long ago wounds have been classified in to four categories called clean, clean-contaminated, contaminated and dirty according to the theoretical number of bacteria that contaminate wounds [4,5]. It has been reported that clean, cleancontaminated, contaminated and dirty wound categories account approximately 1.5-3.9%, 3-4%, 8.5% and 28-40% of wound infection rates respectively [5]. The source of postoperative wound infections can be either endogenously from body flora following surgical manipulation or exogenously from the hospital staffs, other patients and visitors, foods, water, fomites [6-9]. Patients who developed postoperative wound infections are 60% more likely to spend time in an intensive care unit, 5 times more likely to be readmitted and twice more likely to die than patients who don’t have these infections [4].
Most common postoperative wound infection pathogens are Pseudomonas aeruginosa, Staphylococcus aureus, Escherichia coli, Klebsiella species, Proteus species, Citrobacter species and Coagulase negative staphylococci (CONS) [10,11]. The role of these microbes as etiological agents in hospital wound infections is partly because of their ability to survive in the hospital environment by developing resistance to antimicrobials and disinfectants and also their ability to rapidly colonize the body surface of compromised host [9,10]. Rapidly spreading antimicrobial resistance in bacterial populations has made the management and treatment of postoperative wound infections a serious challenge in clinical and surgical practice [12]. Mainly if it is caused by multidrug resistant bacteria it worsens the condition and specifically it has become serious problem in developing countries due to crowding hospital environment, irrational prescription of antimicrobial agents and poor infection prevention program [13]. More over the battle between bacteria and their susceptibility to antibiotics is a problem among public, researchers, clinicians and drug companies who are looking for effective drugs [14].
Materials and Methods
A cross-sectional study was conducted for a period of six months (from March to August 2015) at Tikur Anbessa Specialized Hospital which is the biggest referral hospital of Ethiopia located in the capital city, Addis Ababa. All surgical patients, irrespective of age, operated during the study period that later developed symptoms of post-operative wound infections and who gave informed consent and/or assent to participate on the study were included. However, patients who developed postoperative wound infections later than 30 days after the operation and patients who had infected burn wounds were excluded. A total of 197 study participants were recruited using convenient sampling technique from various surgical wards (orthopedic, ICU, general surgery, pediatrics, obstetrics and gynecology, internal medicine and OPD) of the hospital. The sample size was calculated based on single population proportion from previous study done in Ethiopia where the prevalence was 88.1% (0.881) [11]. Patients were identified from daily operation schedules and those patients who operated during the study period were observed daily for any signs of postoperative wound infection for a total of 30 postoperative days. From eligible participants, patient specific demographic characteristics were collected from the patient card after obtaining informed consent using data collection sheets. Information on potential predicating variables of wound infection and whether wound class was clean, clean contaminated, contaminated and dirty were recorded from the responsible surgeon. Wound swabs were collected aseptically with sterile cotton tipped swabs by the principal investigator, trained nurses and resident doctors. After cleaning skins, the sterile cotton tipped swabs were placed centrally and rolling technique was applied to collect wound samples. All wound swabs were dipped into Stuarts transport media after collection and taken to bacteriology laboratory within 30 minutes for culture and drug susceptibility testing.
Culture and identification
All wound swab specimens were inoculated on blood agar, MacConkey agar and mannitol salt agar and incubated at 37°C for 24 hours aerobically. Identification of bacterial isolates was performed using colony morphology, Gram stain and conventional biochemical tests. Gram positive bacteria were identified at species level using catalase, coagulase, latex agglutination test and Pastorex TM staph-plus (for Staphylococcus aureus identification). Biochemical tests used for identification of Gram negatives to species level were triple sugar iron, indole, citrate, urea, Lysine decarboxylase (LDC), motility and malonat. Klebsiella ozaenae were identified from Klebsiella pneumoniae using malonat biochemical test. Pseudomonas aeruginosa were differentiated from other Pseudomonas species using Pseudomonas Aeruginosa Screen 80 tablet (Rosco, DK- 2630). For all isolated pathogens antimicrobial susceptibility testing was carried out on Muller-Hinton agar using disk diffusion.
Drug susceptibility testing
The disk diffusion method was performed and after 16-18 hours of incubation at 37°C zone of inhibition was measured and interpreted as recommended by the Clinical and Laboratory Standards Institute (CLSI) [15]. Using a sterile wire loop, 3-5 pure colonies were picked from blood agar for Gram positives and MacConkey agar for Gram negatives then emulsified in nutrient broth. Standard inoculums adjusted to 0.5 McFarland standard using McFarland Densitometer was swabbed onto Muller-Hinton agar (dispensed on 100mm plate).
Accordingly detailed CLSI guidelines for each category of Gram positive and Gram negative bacteria, isolates were tested against amikacin (10 μg, Oxoid), amoxicillin (30 μg, BD), amoxicillinclavulanic acid (30 μg, BD), ceftazidime (30 μg, BD), cefotaxime (30 μg, BD), ceftriaxone (30 μg, BD), chloramphenicol (30 μg, BD), clindamycin (2 μg, BD), gentamicin (10 μg, BD), TMP-SXT (1.25 μg+23.75 μg, BD), tetracycline (30 μg, BD), ciprofloxacin (5 μg, BD), penicillin (10 units, BBL), oxacillin (5 μg, BD) and erythromycin (15 μg, BD). Oxacillin susceptibility of Staphylococcus aureus and Coagulase negative Staphylococci was interpreted using 30 μg cefoxitin as a surrogate test. The zone of inhibition was measured to the nearest millimeter and all bacterial isolates were classified as sensitive, intermediate and resistant according to the standardized table supplied by CLSI.
Quality Control
Standard Operating Procedures (SOP) were strictly followed verifying that media meet expiration date and quality control parameters per CLSI (15). Visual inspections of cracks in media or plastic petridishes, unequal fill, hemolysis, evidence of freezing, bubbles, and contamination was done. Quality control was performed to check the quality of medium. Each new lot was quality controlled before use by testing Escherichia coli ATCC 25922 and/or Staphylococcus aureus ATCC25923 standard control strains.
Statistical analysis and interpretation
The data was analyzed using SPSS version 20. The descriptive statistics (mean, percentages or frequency) was calculated. The bi-variant logistic regression analysis was used to see the relation between dependent variables and independent variables. Variables that showed a significant association were selected for further analysis using multiple logistic regression models with a p-value <0.05 considered statistically significant.
Data quality assurance
Socio-demographic characteristics of patients were collected using structured data collection sheets after getting informed consent. Postoperative wound swabs were collected in accordance with SOPs and brought to bacteriology laboratory within 30 minutes for bacteriological analysis. Culture results were recorded carefully before data entry and the data was double checked by a different person before analysis.
Ethics approval and consent to participate
The study was approved by “Department Research and Ethical Review Committee (DRERC)” of the Department of Medical Laboratory Science (MLS/483/15), School of Allied Health Sciences, College of Health Sciences, Addis Ababa University. Written permission letter was also obtained from the study site. The purpose and procedures of the study was explained to the study participants, participants’ parents or guardians within the study period. Those participants who gave informed consent and those children who gave assent and whose parents or guardians gave informed consent were selected and enrolled as the participants of the study. A patient result was communicated to the attending physicians.
Results
Socio-demographic characteristics
One hundred ninety seven (n=197) eligible study participants were investigated during the study period. Of these patients who developed postoperative wound infections 59.9% (n=118/197) of them were males and 40.1% (n=79/197) were females with males to females ratio of 1.49:1. The majority of patients (n=53/197; 27%) and (n=51/197; 25.9%) were between 1-10 and 21-30 years of age as shown in Table 1 and the mean (std. deviation) ages of patients was 24.8(19.2) with age range of 0-85 years. Among all study participants, 14.7% (n=29/197) were out patients while 85.3% (n=178/197) were inpatients. Socio-demographic characteristics of patients have shown in Table 1.
| Variables |
Bacterial culture results (n=197) |
| Positive n (%) |
Negative n (%) |
Total n (%) |
| Gender |
Male |
93(78.8) |
25(38.8) |
118(100) |
| Female |
56(70.9) |
23(29.1) |
79(100) |
| Age in Year |
<10 |
42(79.3) |
11(20.7) |
53(100) |
| Nov-20 |
29(82.9) |
6(17.1) |
35(100) |
| 21-30 |
36(70.6) |
15(19.4) |
51(100) |
| 31-40 |
14(77.8) |
4(22.2) |
18(100) |
| 41-50 |
14(73.7) |
5(26.3) |
19(100) |
| >50 |
14(66.7) |
7(23.3) |
21(100) |
| Patient |
Outpatient |
24(82.8) |
5(17.3) |
29(100) |
| Type |
Inpatient |
125(74.4) |
43(25.6) |
168(100) |
| Wound status |
Dirty |
12(100) |
0(0) |
12(100) |
| Contaminated |
58(98.3) |
1(1.7) |
59(100) |
| Clean contaminated |
63(78.8) |
17(21.2) |
80(100) |
| Clean |
16(34.8) |
30(65.2) |
46(100) |
| Wards (inpatients) |
Orthopedics |
42(82.4) |
9(17.6) |
51(00) |
| Intensive Care Unit |
9(81.8) |
2(18.2) |
11(100) |
| General surgery |
37(75.5) |
12(24.5) |
49(100) |
| Gynecology & obstetrics |
13(68.4) |
6(31.6) |
19(100) |
| Internal Medicine |
4(57.1) |
3(42.9) |
7(100) |
| Pediatrics |
20(64.5) |
11(35.5) |
31(100) |
| Total |
149(75.6) |
48(24.4) |
197(100) |
Table 1: Socio-demographic characteristics of patients who developed postoperative wound infections at TikurAnbessa Specialized Hospital from March to August 2015.
Prevalence of bacterial isolates from postoperative wounds
The overall prevalence of bacterial isolates from postoperative wound infections was 75.6% (n=149/197). Among all bacterial isolates, 62.4% (n=93/149) of the culture positives were from males and 37.6% (n=56/149) were from females. Gram positive and Gram negative isolates constitutes 45.2% (n=76/168) and 54.8% (n=92/168) respectively with Gram positives to Gram negatives ratio of 0.83:1. The frequent bacteria isolated from postoperative wound cultures were Staphylococcus aureus 33.3% (n=56/168). Other isolates were Escherichia coli 14.3% (n=24/168, CONS 11.3% (n=19/168), Acinetobacter spp.10.1% (n=17/168), Klebsiella pneumoniae 8.9% (n=15/168, Pseudomonas spp. 5.3% (n=9/168) and Pseudomonas aeruginosa 4.8% (n=8/168) (Figure 1).
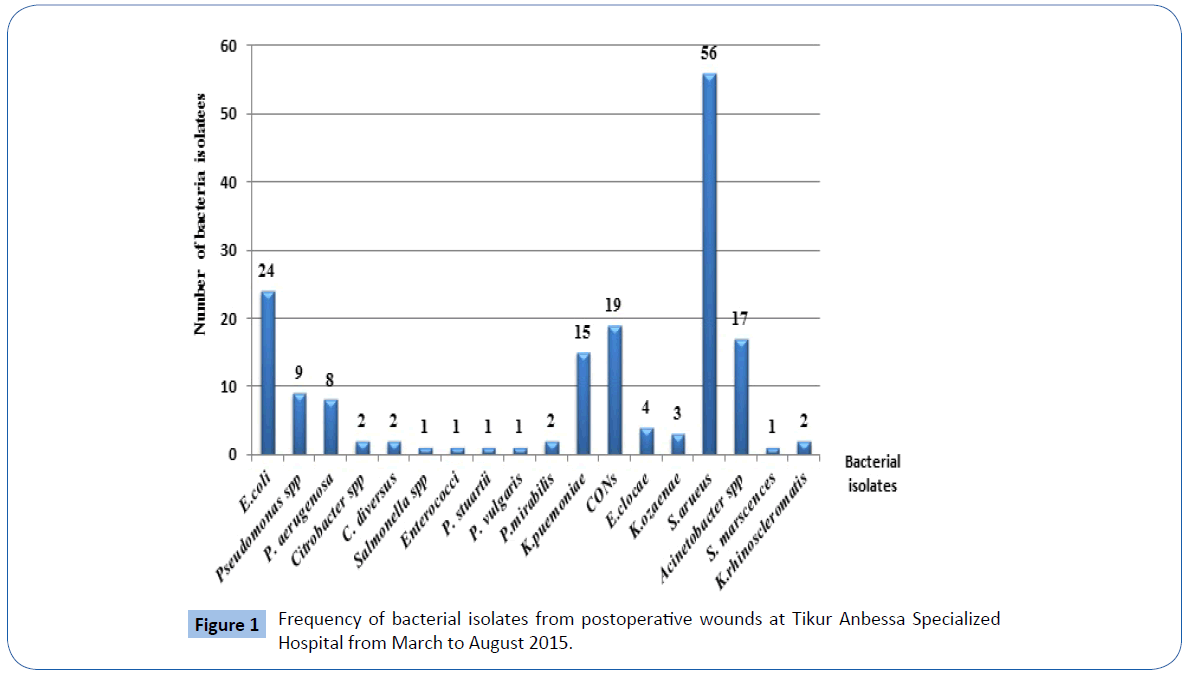
Figure 1: Frequency of bacterial isolates from postoperative wounds at Tikur Anbessa Specialized Hospital from March to August 2015.
The spectrum of post-operative wound infections varied with the age of patients (Table 1). The highest proportion 28.2% (n=42/149) of post-operative wound infections were found in less than 10 years. However, there was no significant association between age of patients and culture results (OR=1.13, 95% CI=0.931-1.377, P = 0.212) (Table 2). In this study, 83.9% (n=125/149) of bacteria were isolated from hospitalized patients while the remaining 16.1% (n=24/149) were from those who attended outpatient department; however there was no significant association between being out patient or inpatient on post-operative wound infection culture results (OR=1.653, 95%CI=0.593-4.597, P = 0.337). Among the different wards, the highest proportion of bacteria 28.2% (n=42/149) were isolated from orthopedics followed by General surgery 24.8% (n=37/149), OPD 16.1% (n=24/149) and pediatrics 13.4% (n=20/149). However, there was no significant association between wards and culture results (OR=1.03, 95%CI=0.309-3.424, P = 0.936).
| Variables |
Culture results (n=197) |
P-value |
OR |
95%CL |
P-value |
AOR |
95%CI |
| Positive n (%) |
Negative n(%) |
|
|
|
|
|
|
| Gender |
Male |
93(78.8) |
25(38.8) |
0.205 |
0.655 |
[0.340-1.262] |
|
|
|
| Female |
56(70.9) |
23(29.1) |
1 |
|
|
|
|
|
| Total |
149(75.6) |
48(24.4) |
|
|
|
|
|
|
| Age in Year |
<10 |
42(79.3) |
11(20.7) |
0.26 |
0.524 |
[0.170-1.612] |
|
|
|
| Nov-20 |
29(82.9) |
6(17.1) |
0.1711 |
0.414 |
[0.117-1.464] |
|
|
|
| 21-30 |
36(70.6) |
15(19.4) |
0.743 |
0.833 |
[0.280-2.476] |
|
|
|
| 31-40 |
14(77.8) |
4(22.2) |
0.445 |
0.571 |
[0.136-2.399] |
|
|
|
| 41-50 |
14(73.7) |
5(26.3) |
0.629 |
0.714 |
[0.182-2.800] |
|
|
|
| <50 |
14(66.7) |
7(23.3) |
1 |
|
|
|
|
|
| Total |
149(75.6) |
48(24.4) |
|
|
|
|
|
|
Patient
Type |
Outpatient |
24(82.8) |
5(17.3) |
1 |
|
|
|
|
|
| Inpatient |
125(74.4) |
43(25.6) |
0.337 |
1.651 |
[0.593-4.597] |
|
|
|
| Total |
149(75.6) |
48(24.4) |
|
|
|
|
|
|
| Wound status |
Dirty |
12(100) |
0(0) |
0.998 |
0 |
0 |
|
|
|
| Contaminated |
58(98.3) |
1(1.7) |
0 |
0.009 |
[0.001-0.073] |
0 |
0.008 |
[0.001-0.063] |
| Clean contaminated |
63(78.8) |
17(21.2) |
0 |
0.144 |
[0.064-0.323] |
0 |
0.133 |
[0.056-0.318] |
| Clean |
16(34.8) |
30(65.2) |
1 |
|
|
1 |
|
|
| Total |
149(75.6) |
48(24.4) |
|
|
|
|
|
|
| OR: Odds Ratio; CI: ConfidenceInterval; AOR: AdjustedOddsRatio; NA: NotApplicable |
Table 2: Association of variables with postoperative wound culture results at Tikur Anbessa Specialized Hospital from March to August 2015.
Among the different wound status of postoperative wound infections clean contaminated wound class yields the highest culture positive results 42.3% (n=63/149) followed by contaminated 25.5% (n=38/149) and clean wounds 10.7% (n=16/149). However, there was no significant association between wound status of the patient and culture results (OR=0.109, 95% CI=0.43-0.274, P = 0.998). Dirty wounds yield 100% (n=12/12) positive culture results though it took the least proportion 6.1% (n=12/197) from all wound status. In our study, mono-microbial isolates were recovered from 88.6% (n=132/149) patients whereas 11.4% (n=17/149) had double microbial infections (Table 3). Highest double microbial infections 52.9% (n=9/17) were showed from orthopedics ward (Table 3). The most double isolated bacteria were Staphylococcus aureus and Pseudomonas spp. 3.5% (n=4/17) combination followed by Staphylococcus aureus and Escherichia coli 17.6% (n=3/17) (Table 4).
| Wards |
Culture results |
| Single infections |
Double infections |
No-Growth |
Total |
| Orthopedics |
33(64.7%) |
9(17.6%) |
9(17.6%) |
51(100%) |
| ICU |
6(54.5%) |
3(27.3%) |
2(18.2%) |
11(100%) |
| General surgery |
35(71.4%) |
2(4.1%) |
12(24.5%) |
49(100%) |
| Gynecology & Obstetrics |
13(68.4%) |
0(0.0%) |
6(31.6%) |
19(100%) |
| Internal medicine |
3(42.9%) |
1(14.3%) |
3(42.9%) |
7(100%) |
| Pediatrics |
19(61.3%) |
1(3.2%) |
11(35.5%) |
31(100%) |
| Out patient |
23(79.3%) |
1(3.4%) |
5(17.2%) |
29(100.0%) |
| Total |
132(67.0%) |
17(8.6%) |
48(24.4%) |
197(100.0%) |
Table 3: Rate of single and double infections from postoperative wounds among different wards at Tikur Anbessa Specialized Hospital from March to
August 2015..
| Double infections |
| Infections |
Frequency |
Percent % |
| K. pneumoniaeand Escherichia coli |
1 |
5.9 |
| Acinetobacter spp. and Escherichia coli |
1 |
5.9 |
| Acinetobacter spp. and Citrobacterdiversus |
1 |
5.9 |
| Staphylococci aureusand Pseudomonas spp. |
4 |
23.5 |
| Staphylococci aureusandAcinetobacter spp. |
1 |
5.9 |
| Staphylococci aureusand Escherichia coli |
3 |
17.6 |
| Staphylococci aureusand K. pneumoniae |
2 |
11.8 |
| P. mirabilis and K. rihinoscleris |
1 |
5.9 |
| Escherichia coli andCitrobacter spp. |
1 |
5.9 |
| Escherichia coli andProvidenciastuartii |
1 |
5.9 |
| Citrobacter spp. and K. rihinoscleris |
1 |
5.9 |
| Total |
17 |
100 |
Table 4: Frequency of double infections from patients who develop postoperative wound infections at Tikur Anbessa Specialized Hospital from March
to August 2015.
Antibiotic resistance patterns
Among the total bacterial isolates (n=168), multidrug resistance (MDR ≥ 2 different classes of drugs) was recorded in 65.5% (n=110/168) of bacterial isolates (Table 5). Antimicrobial resistance level for Gram positive isolates causing postoperative wound infections ranged from 0-100% with an MDR level of 55.3% (n=42/76). Among the Gram positive bacteria, the frequent isolate Staphylococcus aureus 73.7% (n=56/76) showed 44.6% (n=25/56) an MDR level. It showed lowered resistance level for clindamycin (1.8%), chloramphenicol (7.1%) and oxacillin (10.7%) however it demonstrated high level of resistance to penicillin (80.4%) (Table 6). Coagulase negative staphylococci (CONS) showed an MDR level of 84.2% (n=16/19). It showed lower resistance level for sulphamethoxazole-trimethoprim (33.9%), tetracycline (28.6%) and erythromycin (16.1%) however it showed high level of resistance to penicillin (94.7%). It showed better susceptibility for ciprofloxacin (26.3%) and clindamycin (31.6%).
| Organism isolated |
Antimicrobial resistance level in no.(%)of bacterial isolates |
| R2 |
R3 |
R4 |
R5 |
R6 |
R7 |
R8 |
R9 |
R10 |
Total |
| Gram Positive |
| Staphylococci aureus(n=56) |
8(14.3) |
6(10.7) |
3(5.4) |
4(7.1) |
1(1.8) |
1(1.8) |
0(0) |
2(3.6) |
0(0) |
25(44.6) |
| CONS (n=19) |
1(5.3) |
2(10.5) |
2(10.5) |
0(0) |
1(5.3) |
1(5.3) |
4(21.1) |
4(21.1) |
1(5.3) |
16(84.2) |
| Enterococci (n=1) |
0 |
1(100) |
0(0) |
- |
- |
- |
- |
- |
- |
1 |
| Total(n=76) |
9(11.8) |
9(11.8) |
5(6.6) |
4(5.3) |
2(2.6) |
2(2.6) |
4(5.3) |
6(7.9) |
1(1.3) |
42(55.3) |
| Gram Negative |
| K. ozaenae(n=3) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
1(33.3) |
1(33.3) |
1(33.3) |
3(100) |
| P. mirabilis (n=2) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
1(50) |
1(50) |
0(0) |
0(0) |
|
| K. rihinoscleris(n=2) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
1(50) |
1(50) |
0(0) |
0(0) |
|
| S.marscensence(n=1) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
1(100) |
0(0) |
0(0) |
|
| Citrobacter spp. (n=2) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
1(50) |
0(0) |
0(0) |
|
| Citrobacterdiversus(n=2) |
0(0) |
0(0) |
0(0) |
0(0) |
1(50) |
0(0) |
0(0) |
1(50) |
|
2(100) |
| Providenciastuartii(n=1) |
0(0) |
0(0) |
0(0) |
1(100) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
1(100) |
| Salmonella spp. (n=1) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
1(100) |
0(0) |
0(0) |
1(100) |
| K. pneumoniae(n=15) |
0(0) |
0(0) |
2(13.3) |
4(26.7) |
0(0) |
2(13.3) |
4(26.7) |
2(13.3) |
0(0) |
14(93.3) |
| P. aerogens(n=8) |
1(12.5) |
1(12.5) |
0(0) |
0(0) |
- |
- |
- |
- |
|
2(25) |
| Pseudomonas spp. (n=9) |
3(33.3) |
3(33.3) |
0(0) |
0(0) |
0(0) |
0(0) |
- |
- |
|
6(66.7) |
| Acinetobacter spp. (n=17) |
1(5.9) |
1(5.9) |
1(5.9) |
4(23.5) |
0(0) |
0(0) |
- |
- |
|
7(41.2) |
| Escherichia coli (n=24) |
1(4.2) |
2(8.3) |
2(8.3) |
3(12.5) |
2(8.3) |
2(8.3) |
6(25) |
2(8.3) |
1(4.2) |
21(87.5) |
| P. vulgaris (n=1) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
0(0) |
1(100) |
0(0) |
0(0) |
1(100) |
| E. cloacae (n=4) |
1(25) |
1(25) |
1(25) |
0(0) |
0(0) |
0(0) |
0(0) |
1(25) |
0(0) |
4(100) |
| Total (n=92) |
7(7.6) |
8(8.7) |
6(6.5) |
12(13.0) |
3(3.3) |
6(6.5) |
17(18.5) |
7(7.6) |
2(2.2) |
68(73.9) |
| CNS: Coagulase negativestaphylococci; R2, R3, R4, R5, R6, R7, R8, R9, R 10-Resistant totwo, three, four, five, six, seven, eight, nine and ten antimicrobialsrespectively. |
Table 5: Multidrug resistance pattern of bacteria isolated from postoperative wounds at Tikur Anbessa Specialized Hospital from March
to August 2015.
| Organism isolated |
|
Antimicrobial resistance level in no.(%)of bacterial isolates from postoperative wounds |
| |
AML |
AMC |
SXT |
PEN |
CLD |
VA |
OX |
ERT |
GE |
CTX |
CAZ |
C |
TE |
AN |
CIP |
CRO |
| S. aureus (n=56) |
NT |
9(16.1) |
19(33.9) |
45(80.4) |
1(1.8) |
NA |
6(10.7) |
9(16.1) |
7(12.5) |
NA |
NA |
4(7.1) |
16(28.6) |
NA |
7(12..5) |
NT |
| K. ozaenae(n=3) |
3(100) |
3(100) |
3(100) |
NA |
NA |
NA |
NA |
NA |
3(100) |
3(100) |
3(100) |
2(66.7) |
3(100) |
0(0) |
2(66.7) |
3(100) |
| P. mirabilis (n=2) |
2(100) |
2(100) |
2(100) |
NA |
NA |
NA |
NA |
NA |
2(100) |
1(50) |
0(0) |
2(100) |
2(100) |
0(0) |
0(0) |
1(50) |
| K. rihinoscleris (n=2) |
2(100) |
2(100) |
2(100) |
NA |
NA |
NA |
NA |
NA |
0(0) |
2(100) |
2(100) |
1(50) |
2(100) |
0(0) |
2(100) |
2(100) |
| S. marscensence (n=1) |
1(100) |
1(100) |
1(100) |
NA |
NA |
NA |
NA |
NA |
1(100) |
1(100) |
1(100) |
1(100) |
1(100) |
0(0) |
0(0) |
1(100) |
| Citrobacter spp. (n=2) |
1(50) |
1(50) |
1(50) |
NA |
NA |
NA |
NA |
NA |
1(50) |
1(50) |
1(50) |
0(100) |
1(50) |
0(0) |
1(50) |
1(50) |
| C. diversus (n=2) |
2(100) |
2(100) |
1(50) |
NA |
NA |
NA |
NA |
NA |
2(100) |
2(100) |
2(100) |
1(50) |
1(50) |
0(0) |
2(100) |
2(100) |
| Enterococci (n=1) |
NA |
NA |
NA |
1(100) |
NA |
0(0) |
NA |
NA |
NA |
NA |
NA |
1(00) |
1(100) |
NA |
NA |
NA |
| P. stuartii (n=1) |
1(100) |
1(100) |
1(100) |
NA |
NA |
NA |
NA |
NA |
0(0) |
1(100) |
0(0) |
1(100) |
0(0) |
0(0) |
0(0) |
1(100) |
| Salmonella spp. (n=1) |
1(100) |
1(100) |
1(100) |
NA |
NA |
NA |
NA |
NA |
1(100) |
1(100) |
1(100) |
1(100) |
1(100) |
0(0) |
0(0) |
1(100) |
| K. pneumoniae (n=15) |
14(93.3) |
12(80) |
13(86.7) |
NA |
NA |
NA |
NA |
NA |
9(100) |
13(86.7) |
9(60) |
7(46.7) |
9(60) |
0(0) |
4(26.7) |
13(86.70) |
| P. aerogens (n=8) |
8(100) |
7(87.5) |
7(87.5) |
NA |
NA |
NA |
NA |
NA |
2(25.0) |
NA |
1(12.5 |
3(37.5) |
8(100) |
1(12.5) |
1(12.7) |
NA |
| Pseudomonas spp.(n=9) |
9(100) |
8(88.9) |
8(88.9) |
NA |
NA |
NA |
NA |
NA |
7(77.8) |
NA |
6(66.7) |
7(77.8) |
9(100) |
0(0) |
3(33.3) |
NA |
| Acinetobacter spp.(n=17) |
17(100) |
15(88.2) |
15(88.2) |
NA |
NA |
NA |
NA |
NA |
12(70.6) |
17(100) |
16(94.1) |
13(76.5) |
14(82.4) |
0(0) |
12(70.6) |
17(100) |
| Escherichia coli (n=24) |
20(83.3) |
14(58.3) |
15(62.5) |
NA |
NA |
NA |
NA |
NA |
7(29.2) |
15(62.5) |
14(58.3) |
4(16.7) |
15(62.5) |
0(0) |
9(37.5) |
17(70.8) |
| CONS (n=19) |
NT |
15(78.9) |
14(73.7) |
18(94.7) |
6(31.6) |
NA |
10(52.6) |
11(57.9) |
12(63.2 |
NA |
NA |
9(47.4) |
12(63.2) |
NA |
5(26.3) |
NA |
| P. vulgaris (n=1) |
1(100) |
1(100) |
1(0) |
NA |
NA |
NA |
NA |
NA |
1(100) |
1(100) |
1(100) |
0(0) |
1(100) |
0(0) |
1(100) |
1(100) |
| E. cloacae (n=4) |
4(100) |
2(50) |
1(25) |
NA |
NA |
NA |
NA |
NA |
1(25) |
4(100) |
4(100) |
1(25) |
1(25) |
0(0) |
1(25) |
4(100) |
| Total(n=168) |
86(93.5) |
96(57.5) |
105(62.9) |
64(84.2) |
7(9.3) |
0(0) |
16(21.3) |
20(26.7) |
68(40.7) |
62(82.7) |
49(53.3) |
57(33.9) |
97(57.7) |
1(1.1) |
50(29.8) |
64(85.3) |
| CNS: Coagulase Negative Staphylococci, NA: Not Applicable; NT: Not Tested, AML: Amoxicillin, AmC: Amoxicillin-Clavulanic Acid, AN: Amikacin, SXT: Sulphamethoxazole-Trimethoprim; C: Chloramphenicol, CTX: Cefotaxime, CAZ: Ceftazidme, CRO: Ceftriaxone, GM: Gentamicin; TE: Eetracycline; CIP: Ciprofloxacin; OX: Oxacillin; ERY: Erythromycin; CLD: Clindamycin; VA: Vancomycin; PEN: Penicillin |
Table 6: Antimicrobial resistance levels of bacterial isolates from postoperative wounds at Tikur Anbessa Specialized Hospital from March to August 2015.
In the same manner to gram positive bacteria, the resistance patterns of Gram negative organisms causing postoperative wound infections were ranged from 0-100% and they showed 73.9% (n=68/92) of an MDR level. Almost all gram negative bacterial isolates showed greater than 90% level of resistance for amoxicillin and almost all isolates were sensitive for amikacin (>95%). Among the Gram negative bacteria, the frequent isolates, Escherichia coli 24.1% (n=24/92), showed 87.5% (n=21/24) of MDR level which demonstrated highest level of resistance to amoxicillin (90%). It showed lower resistance level for chloramphenicol (16.7%) and gentamicin (29.2%). Among the antibiotics tested, amoxicillin (93.5%), ceftriaxone (85.3%), penicillin (84.5%) and cefotaxime (82.7%) were least effective. Clindamycin (7.9%) for Gram positives and amikacin (1.1%) for Gram negative isolates were the most effective drugs.
Discussion
Prevalence of bacterial isolates among postoperative wound infections
The overall prevalence of bacterial isolates from postoperative wounds with clinical suspicion of wound infections was 75.6% (n=149/197) which was in line with previous studies done in Ethiopia which showed 75% and 71.1% of prevalence [16,17]. On the contrary, lower rates of isolation (11.4%, 10.9%) were reported from other localities of Ethiopia [18,19]. However, our finding was relatively lower than other studies done in Ethiopia (92%, 87.3%) [20,21] and India (96%) [22]. The possible explanation for the difference could be varying bacterial etiology and infection prevention practices in diverse geographical settings and at different sampling times [17]. The effect of antimicrobials used for surgical prophylaxis, that the infection is already resolving, antiseptics used for cleaning the wounds, bacteria fail to grow due to their fastidious nature, samples only containing already dead bacteria or wound infections caused by unidentifiable pathogens by the methods used could be main factors for the absence of bacterial growth in samples collected from surgical wounds with clinical signs of infections [19,20].
The incidence of wound infection was higher in males 62.4% (n=93/149) than in females 37.6% (n=56/149). However, there was no significant association between sex of patient and culture results (OR=0.65, 95% CI=0.340-1.262, P=0.205). This showed agreement with studies done in different parts of Ethiopia and other countries [17,21,23]. This might be explained by the fact that traditionally, in Ethiopia, males are predominantly involved in occupations such as farming, construction works, transportation and industry works where they are at higher risk of trauma [21] and exposed to the outside environment than females [24].
The spectrum of post-operative wound infections varied with the age of patients (Table 5) where 28.2% (n=42/149) of postoperative wound infections occurred in less than 10 years in contrary to other study [24]. However, there was no statistical significant association between age of patient and culture results (OR=1.13, 95% CI=0.931-1.377, P=0.212). It showed disagreement with a study done in Ethiopia which reported that age of the patient showed statically significant association with culture results [25]. In this study 83.9% (n=125/149) of bacteria were isolated from hospitalized patients while the remaining 16.1% (n=24/149) were from those who attended outpatient departments; however there was no significant association between being out patient or inpatient on post-operative wound infection culture results (OR=1.653, 95%CI=0.593-4.597, P = 0.337). It was a similar finding with a previous study done in Ethiopia [17].
Among the different wards, the highest proportion of bacteria 28.2% (n=42/149) were isolated from orthopedics which showed similarity with a previous study [16]. However, there was no statistical significant association between different wards and culture results (OR=1.03, 95%CI=0.309-3.424, P=0.936). While General surgery 24.8% (n=37/149) and OPD 16.1% (n=24/149) took the second and third rank of bacteria proportion, pediatrics 13.4% (n=20/149), gynecology and obstetrics 8.7% (n=13/149) and internal medicine 2.7% (n=4/149) followed them. It also showed similarity with other study done in Nepal [24]. This could be due to that in orthopedic wards since patients require longer hospitalization time to recover from their bone cases, they become prone for post-operative wound infections [24].
In our study, among the different classes of postoperative wound infections clean contaminated wound showed highest culture positive results 42.3% (n=63/149). Multivariate regression analysis showed a statistical significant association between wound status of the patient and culture results (OR=0.144, 95% CI=0.0640-0.323, P=0.00). It showed agreement with other study done in Ethiopia [17]. This high rate of infections could be probably because of profound influence of endogenous contamination during the time of operation [17]. Contaminated wounds 58.9% (n=38/149) and clean wounds 10.7% (n=16/149) took the second and third culture positive ranks following to clean contaminated wounds. In the case of dirty wounds, it yields 100% (n=12/12) positive culture results though it took the least proportion from all wound status [16].
Double bacterial infections, among culture positive post-surgical wound infected patients, was seen in 11.4% (n=17/149) of patients. The rate of double pathogens in the current study was comparable with a previous study done in South West Ethiopia [21] however it was a lower finding compared to another study conducted in Southern Ethiopia (20.1%) [17]. The leading double infection was caused by Staphylococcus aureus in combination with Pseudomonas species was also found in another study from Ethiopia [25].
In our study, Gram negatives 54.8% (n=92/168) were isolated more commonly than Gram positive isolates 45.2% (n=76/168) which showed similarity with other studies [24,26]. The most frequent bacteria isolated in this study were Staphylococcus aureus 33.3% (n=56/168). It showed similarity with other studies done throughout the world [17,26,27]. The predominance (33.3%) of Staphylococcus aureus infection seen in this study could be associated with endogenous source as the organism colonizes the skin that lead access to deep sites during surgery [16]. This organism may also be transmitted from the environment, surgical instruments or contaminated hands of the health professionals [19]. A study done in Nepal found that Pseudomonas spp. were the most frequently isolated bacteria from postoperative wound infections [24]. The possible reason for these varied findings with our study could be due to populations; different surgical procedures as well as timing of specimen collections [24].
The second most common bacteria isolated in this study was Escherichia coli 14.3% (n=24/168) that showed similarity with a study done in Nepal (17.5%) [24]. Endogenous contamination from bowel flora of patients could explain this finding [19]. Other bacterial species found in this study were Coagulase negative Staphylococci 11.3% (n=19/168), Acinetobacter spp. 10.1% (n=17/168), Klebsiella pneumoniae 8.9% (n=15/168, Pseudomonas spp. 5.3% (n=9/168) and Pseudomonas aeruginosa 4.8% (n=8/168).
Antimicrobial resistance patterns of bacterial isolates
The overall multidrug resistance level of all bacterial isolates was 65.5% (n=110/168). This finding was in line with the finding of a study done in other part of Ethiopia where the MDR level was 65.2% [25]. On the other hand, our finding was a lowered finding as compared to what has been recorded in South West Ethiopia which was 85.2% [21]. In our study, Gram positive and Gram negative bacteria showed 55.3% (n=42/76) and 73.9% (n=68/92) MDR level respectively. This high rate of antibiotic resistance might reflect inappropriate use of antibiotic, lack of laboratory diagnostic tests for appropriate antibiotic selection, unavailability of guideline for the selection of antibiotics, unskilled practitioners, expired antibiotics, self-medication, counterfeit drugs, or inadequate hospital control measures [28].
The most frequently isolated pathogens, Staphylococcus aureus, showed 44.6% (n=25/56) of an MDR level which disagreed with previous studies done in Ethiopia [19,20] where Staphylococcus aureus showed 100% MDR level. The difference could be due to the difference in prescribing this antibiotic for the treatment hospital to hospital [16]. It demonstrated high level of resistance to penicillin (80.4%) which was consistent with a study done in other part of Ethiopia [21]. It showed lower resistance level for sulphamethoxazole-trimethoprim (33.9%), tetracycline (28.6%), erythromycin (16.1%), clindamycin (1.8%), chloramphenicol (7.1%) and oxacillin (10.7%) as compared to other tested drugs.
Among the Gram negative bacteria, the predominant isolates, Escherichia coli 24.1% (n=24/92), showed 87.5% (n=21/24) of multidrug resistance level, It demonstrated high level of resistance to amoxicillin (90%) and a lowered resistance level for chloramphenicol (16.7%) and gentamicin (29.2%). This finding showed disagreement with a study done in Ethiopia where Escherichia coli isolates showed highest resistant level to ampicillin (96.6%), tetracycline (79%), chloramphenicol (65.5%), ceftriaxone (62%), sulphamethoxazole-trimethoprim (55%) and gentamicin (51.7%). The difference could be due to the difference in prescribing these antibiotics for the treatment of the bacteria from hospital to hospital [16]. In our study, amoxicillin (93.5%), ceftriaxone (85.3%), penicillin (84.2%) and cefotaxime (82.7%) were least effective drugs.
Conclusion
The overall 75.6% (n=149/197) prevalence of bacterial isolates from postoperative wound infections was high and the most frequent isolated bacteria were Staphylococcus aureus and Escherichia coli. Although complete eradication of postoperative wound infections is not possible, proper precautions should be taken to minimize the occurrence by strictly adhering aseptic surgical procedures and proper management of wounds. The choice of drugs for the treatment of bacterial isolates from postoperative wound infections was quit narrow especially for bacterial strains which were resistant to most classes of antibiotics which have been used previously. To prevent further emergence and spread of MDR bacterial pathogens, rational use of antibiotics and regular monitoring of antimicrobial resistance patterns are essential and mandatory.
Competing of interest
The Authors declared that no competing of interests.
Acknowledgments
We greatly appreciate Addis Ababa University for supporting this study. We are also grateful to the Department of Clinical Laboratory Sciences to give ethical clearance for this study. Our deep gratitude goes to those study participants and also parents and guardians who gave us their willingness to participate in this study by giving written consent form.
20137
References
- Gaynes RP, Culver DH, Horan TC, Edwards JR, Richards C, et al. (2001) Surgicalsiteinfection(SSI)ratesintheUnitedStates,1992-1998:TheNationalNosocomialInfectionsSurveillanceSystembasic SSI riskindex. ClinInfectDis 33: S69-S77.
- PryorKO, Fahey TJ, Lien CA, Goldstein PA (2004) Surgicalsiteinfection and theroutine. Use of perioperativehyperoxia in a general surgicalpopulation: Arandomizedcontrolled trial.JAMA 29: 79-87.
- Lilani SP, Jangale N, Chowdhary A, Daver GB (2005) Surgicalsiteinfection in clean and clean-contaminated cases. Indian J MedMicrobiol 23:249-252.
- GraySH,HawnMT (2007)Preventionofsurgicalsiteinfections. Hospital physician. RevObstetGynecol43: 41.
- Tietjen L, Bossemeyer D, McIntosh N (2003) Infectionprevention: Guidelinesforhealthcarefacilitieswithlimitedresources. USA.
- Nichols RL (2001) Preventingsurgicalsiteinfections: Asurgeon'sperspective. EmergInfectDis 7: 220.
- Endalafer N, Gebre-Selassie S, Kotiso B(2010)Nosocomial bacterialinfections in a tertiary Hospitalin Ethiopia. J InfectPrevent. 12: 38-43.
- Nelson J, Bivens A, Shinn A, Wanzer L, Kasper C (2006) Microbial flora onoperatingroomtelephones. AORN J 83:607-626.
- KramerA,SchwebkeI,KampfG (2006)Howlongdonosocomialpathogenspersistoninanimatesurfaces? A systematicreview. BMC InfectDis 6:130.
- AnguzuJ,OlilaD (2007)Drugsensitivitypatternsofbacterialisolatesfromsepticpost-operativewounds in a regional referral hospital in Uganda. AfrHealthSci 7: 148-154.
- GelawA,Gebre-SelassieS,TirunehM,MathiosE,YifruS (2014) Isolationofbacterialpathogensfrompatientswithpostoperativesurgicalsiteinfectionsandpossiblesourcesof infections at theUniversity of Gondar Hospital, NorthwestEthiopia. J EnvironOccupSci 3:103-108.
- Isibor JO, Oseni A, Eyaufe A, Turay A (2008) Incidence of aerobic bacteria and Candidaalbicansinpost-operativewoundinfections.Afr J Microbiol Res 2:288-291.
- MesseleG,WoldemedhinY,DemissieM,MamoK,GeyidA (2009)Commoncausesof nosocomial infections and theirsusceptibilitypatterns in twohospitals in Addis Ababa. Ethiop J HealthBiomedSci 2:3-8.
- Biadglegne F, Abera B, Alem A, Anagaw B (2009) Bacterialisolatesfromwoundinfection and theirantimicrobialsusceptibilitypattern in FelegeHiwotreferral Hospital North West Ethiopia. Ethiop J HealthSci2017: 1-10.
- CLSI (2014) Performance StandardsforAntimicrobial Disk SusceptibilityTests; Twenty-ThirdInformationalSupplement. CLSI Document 33: M100-S23.
- Mengesha RE, Kasa BG-S, Saravanan M, Berhe DF, Wasihun AG(2014)Aerobic bacteria in post-surgicalwoundinfectionsandpatternoftheirantimicrobialsusceptibilityinAyderTeaching and Referral Hospital, Mekelle, Ethiopia. BMC Res Notes 7: 575.
- Dessalegn L, Shimelis T, Tadesse E, Gebre-selassie S (2014) Aerobic bacterialisolatesfrom post-surgicalwoundandtheirantimicrobialsusceptibilitypattern:Ahospitalbasedcross-sectionalstudy. E3 J MedRes 3: 18-23.
- Amenu D, Belachew T, Araya F (2011) Surgicalsiteinfectionrate and riskfactorsamongobstetric cases of JimmaUniversitySpecialized Hospital, SouthwestEthiopia. Ethiop J HealthSci 2:91-100.
- MuluW,KibruG,BeyeneG,DamtieM (2012) Postoperativenosocomialinfectionsand antimicrobialresistancepattern of bacteria isolatesamongpatientsadmitted at FelegeHiwotReferral Hospital, Bahirdar, Ethiopia. Ethiopia J HealthSci 22:7-18.
- GutaM,AragawK,MeridY (2014) Bacteriafrominfectedsurgicalwoundsandtheirantimicrobialresistance in HawassaUniversityReferralTeaching Hospital, SouthernEthiopia. African JMicrobiol Res 8: 1118-1124.
- Mama M, Abdissa A, Sewunet T (2014) Antimicrobialsusceptibilitypattern of bacterialisolatesfromwoundinfection and theirsensitivitytoalternativetopicalagents at JimmaUniversitySpecialized Hospital, South-West Ethiopia. Ann ClinMicrobiolAntimicrob 13:14.
- Rao R, Sumathi S, Anuradha K, Venkatesh D, Krishna S (2013) Bacteriology of postoperativewoundinfections. Int J PharmBiomed Res 4: 72-76.
- NobandeganiZinat M, DoulatabadShahla N, Masoumeh R, Ardeshir A (2011) Surgicalsiteinfectionincidenceafter a clean-contaminatedsurgery in YasujShahidBeheshti hospital, Iran. InvestEducEnferm 29:435-441.
- Amatya J, Rijal M, Baidya R (2015) Bacteriologicalstudy of thepostoperativewoundsamplesandantibioticsusceptibilitypatternoftheisolatesinB&BHospital.JSMMicrobio 3: 1019.
- AzeneMK,BeyeneBA (2011) BacteriologyandantibiogramofpathogensfromwoundinfectionsatDessieLaboratory,NorthEastEthiopia.Tanzan J Health Res 13: 68-74.
- BhattCP,BaidyaR,KarkiP,ShahRK,MiyaR, etal. (2014)Multidrugresistancebacterialisolates of surgicalsiteinfection. Open J MedMicrobiol4: 203-209.
- Mwambete K, Rugemalila D (2015) Antibioticresistanceprofiles of bacteria isolatedfromsurgicalwounds in tertiaryhospitals, Tanzania. Int J CurrMicrobiol App Sci 4: 448-455.
- Fehr J, Hatz C, Soka I, Kibatala P, Urassa H, et al. (2006) Riskfactorsforsurgicalsiteinfection in a Tanzaniandistrict hospital: a challengeforthetraditionalNational Nosocomial InfectionsSurveillancesystemindex. Infect Control HospEpidemiol 27: 1401-1404.







