Foozieh Arabzadeh1, Fatemeh Aeini2, Fatemeh Keshavarzi2* and Pezhman Karami3
1Department of Biology, Kurdistan Science and Research Branch, Islamic Azad University, Sanandaj, Iran
2Department of Biology, Sanandaj Branch, Islamic Azad University, Sanandaj, Iran
3Faculty of Medicine, Department of Microbiology, Hamedan University of Medical Science, Hamedan, Iran
- *Corresponding Author:
- Dr. Fatemeh Keshavarzi
Ph.D, Assistant Professor
Department of Biology, Sanandaj Branch
Islamic Azad University, Sanandaj, Iran
Tel: +98 91 8370 4918
E-mail: gol.keshavarzi@gmail.com
Received date: October 20, 2018; Accepted date: October 26, 2018; Published date: October 30, 2018
Citation: Arabzadeh F, Aeini F, Keshavarzi F, Karami P (2018) Resistance to Tetracycline and Vancomycin of Staphylococcus aureus Isolates from Sanandaj Patients by Molecular Genotyping. Ann Clin Lab Res Vol.6 No.4:260. doi:10.21767/2386-5180.100260
Keywords
Staphylococcus aureus; Antibiotic resistance; Tetracycline; Vancomycin; Sanandaj
Introduction
During the last decade the prevalence of staphylococcus aureus species have been increasing worldwide. Staphylococcus can grow in different environmental conditions but the best condition for their growth is 30 to 37°C. These bacteria are resistant and they can colonize on the skin and mucous membranes. Staphylococcus aureus species are one of the first known human pathogens. For the first time in 1880 and 1882 in laboratory and clinical studies, the disease caused by staphylococcus and its role in the growth of abscess was explained. Nowadays after 100 years, this organism is considered as one of the dangerous human pathogens. In general this bacterium is really important due to its tenacity destruction, potential power and increasing resistance to antibiotics [1]. Staphylococcus includes at least 40 species. Most of staphylococcus species are harmless and they reside on the skin and the mucosa of the human body and other animals. In the USA, staphylococcus aureus is responsible for the half of positive blood cultures among patients in ICU and it is one of the most important factors of pneumonia in these units [1,2]. The most population of staphylococcus is on the skin in human beings, especially in groin, armpit, perianal region and anterior nasal apertures and it can cause infection in all age groups. In terms of the place of getting infection, they can be divided into two groups: community acquired and hospital acquired. The content of cytosine +guanine in their DNAs is 30% to 40%. Staphylococcus species are anaerobic and they grow in the presence of bile salts and all of them are catalase-positive unlike streptococci. One of the prominent features in classifying the staphylococcus is producing coagulase enzyme [3]. Now 6 species of staphylococcus are coagulase-positive include staphylococcus aureus, staphylococcus delphini, staphylococcus hyicus, staphylococcus intermedius, staphylococcus lutrae, staphylococcus pseuditermedius and staphylococcus schleiferi the subspecies of coagulans. The most important species of coagulasenegative is staphylococcus epidermiditis which resides on the skin of human beings as a symbiotic (coexist). In the patients whose immune systems have been suppressed or those who use counters vessel, these bacteria can cause severe infections. Staphylococcus saprophyticus is another coalasenegative species which is considered as a normal vaginal flora in women. This bacterium has a role in female genital tract infections among sexually active young women [4,5]. According to what was said, the purpose of this research is to study the prevalence and genetic basis of the resistance to antibiotics such as tetracycline and vancomycin in staphylococcus aureus samples in Sanandaj. The findings of this research may be helpful to choose the medicine and for better treatment in the region.
Materials and Methods
Analysis method
Samples: 150 staphylococcus aureus bacterial strains are collected from medical diagnostic laboratories in Sanandaj and Gram stain was done on all the samples to confirm that the bacterium is positive. After confirming Gram-positive bacteria, stock was provided from all collected samples which were on the Mueller-Hinton and blood agar. The biochemical differentiating tests to confirm the staphylococcus aureus included catalase, coagulase, DNAase, novobiocin sensitivity test and mannitol salt agar.
McFarland turbidity and antibogram test
BaSo4 was used according to the McFarland standard to standardize the inoculation concentration for the antimicrobial susceptibility test. The McFarland standard of BaSo4 was prepared as following: half ml of (BaCl2) 0.048 mol/l (W/V BaCl2. 2H2O% 1/175) was added to 99/5 ml sulfuric acid 0.18 mol/l (%1 V/V) and the suspension was prepared by constant stirring. The correct density of standard turbidity was determined through measuring the absorption in a spectrophotometer at optical path length of 1 cm then the amount of 4-6 ml was poured into the screw-cap tubes, the same size as bacterial suspension tubes. After isolating the bacteria, some colonies of bacteria were removed by once (not loop) and they were dissolved in physiology serum. After preparing the homogeneous solution, it is stirred with a sterile swab then after rinsing off, the swab is transferred to the Mueller-Hinton medium and it is cultivated as meadow. Based on the FAO’s table, if the bright halo of staphylococcus aureus is 14 mm or less, it is considered as a resistant strain and if this halo is 15 to 18 mm, it is considered as intermediate resistant strain and if the mentioned halo is 19 mm or more, the strain is considered as susceptible. For vancomycin, if the halo is 11 mm or less, the strain is resistant, if this halo is 11 to 14 mm, the strain is intermediate resistant and if the halo is 15 mm or more, it is susceptible.
DNA extraction and PCR
Genome of those colonies which were cultivated in the selective medium of cultures and those which biochemical tests were done on them and were determined as the staphylococcus aureus bacteria, were extracted by DNA extraction kit of Gram-positive bacteria (CinnaGen company). Then PCR was done by the primers of Table 1. Target genes in thermocycler were done through a plan consists of the initial denaturation in 94°C for 5 minutes and then 30 cycles of denaturation in 92°C for 30 seconds, annealing in 50 to 60°C for 30 seconds and extension in 72°C for 1 minute and then the final extension in 72°C for 10 minutes. Size marker of 100 (bp) was used to determine the size of bands in this research.
| Genes |
Primer’s Sequence of Genes |
| Tet (M)-R |
5-TCCGACTATTTAGACGACGG-3 |
| Tet (M)-F |
5-AGTTTTAGCTCATGTTGATG-3 |
| Tet (K)-R |
5-GTAGTGACAATAAACCTCCTA-3 |
| Tet (K)-F |
5-GTAGCGACAATAGGTAATAGT-3 |
| Tet (O)-R |
5-TCCCACTGTTCCATATCGTCA-3 |
| Tet (O)-F |
5-AACTTAGGCATTCTGGCTCAC-3 |
| Tet (L)-R |
5-AACCAGCCAACTAATGACAAGAT-3 |
| Tet (L)-F |
5-ATAAATTGTTTCGGGTCGGTAAT |
| Van (A)-R |
5-TCACCCCTTTAACGCTAATA-3 |
| Van (A)-F |
5-ATGAATAGAATAAAAGTTGC-3 |
Table 1: Genes responsible for resistance and their primerâ€Âââ€Å¾Â¢s sequence.
Results
In this research, first 192 samples of bacteria were collected then 150 samples of collected bacteria were used as staphylococcus aureus through biochemical and differentiation tests.
Antibiotic susceptibility test results
12% of staphylococcus aureus were resistant to vancomycin (18 out of 150 samples), 52% of samples were intermediate resistant (78 out of 150 samples) and 36% of them were susceptible to vancomycin (54 out of 150 samples) (Figure 1).
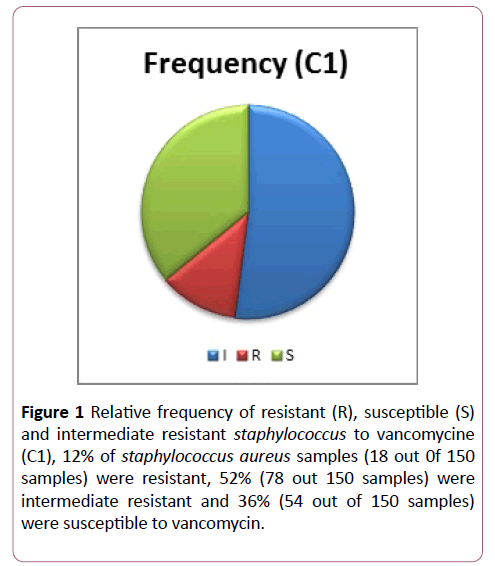
Figure 1: Relative frequency of resistant (R), susceptible (S) and intermediate resistant staphylococcus to vancomycine (C1), 12% of staphylococcus aureus samples (18 out 0f 150 samples) were resistant, 52% (78 out 150 samples) were intermediate resistant and 36% (54 out of 150 samples) were susceptible to vancomycin.
In addition, according to the antibiotics test results, 61/33% of staphylococcus aureus were resistant to tetracycline (92 out of 150 samples), 26% were intermediate resistant (39 out of 150 samples) and 12/667% of them were susceptible to tetracycline (19 out of 150 samples) (Figure 2).
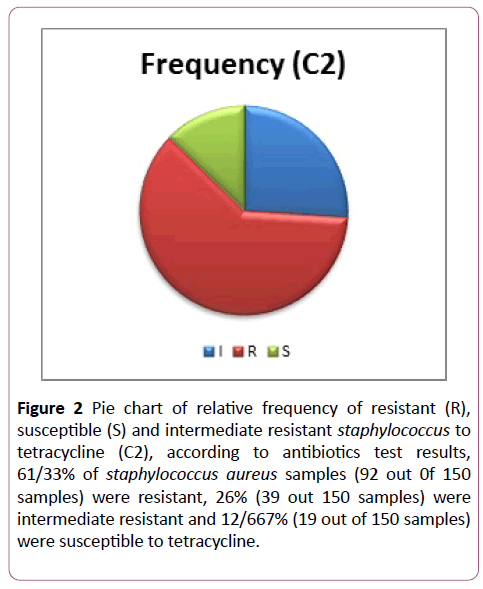
Figure 2: Pie chart of relative frequency of resistant (R), susceptible (S) and intermediate resistant staphylococcus to tetracycline (C2), according to antibiotics test results, 61/33% of staphylococcus aureus samples (92 out 0f 150 samples) were resistant, 26% (39 out 150 samples) were intermediate resistant and 12/667% (19 out of 150 samples) were susceptible to tetracycline.
DNA extraction and PCR results of staphylococcus aureus
According to the PCR results, 47/33% had tet (K) gene (71 out of 150 samples of staphylococcus aureus). Also 60/86% had tet (K) gene (56 out of 92 samples of resistant staphylococcus aureus to tetracycline) and it was observed in 15 samples of intermediate resistant samples (38/46% of intermediate resistant bacteria). Van (A) gene was not observed in this research. Also 21/33% had tet (M) gene (32 out of 150 samples of staphylococcus aureus). Also 30% had tet (M) gene (28 out of 92 samples of tetracycline resistant staphylococcus aureus). Four samples of tetracycline intermediate resistant staphylococcus aureus had this gene. In addition, 21 out of 32 tet (M) staphylococcus aureus samples had tet (K) gene as well (65/62%). Foremore 0/66% (1 out of 150 staphylococcus aureus samples) and 1/8% of tetracycline resistant staphylococcus aureus had tet (O) gene (1 staphylococcus aureus bacterium of 92 bacteria). This sample had tet (K) gene as well. Results showed that 3/33% (5 out of 150 staphylococcus aureus samples) and 5/43% of tetracycline resistant staphylococcus aureus samples had tet (L) gene (5 staphylococcus aureus bacteria of 92 bacteria). It should be mentioned that 3 samples of 5 staphylococcus samples had both tet (L) and tet (K) gene (60% of tet (L) samples) and one sample of these 5 samples had tet (M) (20%). Only one sample out of 150 staphylococcus aureus samples had three genes of tet (K), tet (M) and tet (L) (0/66%).
Discussion
According to National Nosocomial Infection Surveillance Committee, 75% of negative-coagulase staphylococcus and 47% of positive-coagulase strains which were isolated from intensive care unit in December of 2000 were resistant to methicillin, so vancomycin is the first selective drug in the treatment of infections [6]. Vancomycin is widely used in the treatment of infections caused by methicillin resistant staphylococcus aureus which leads to the increasing vancomycin resistant and the spread of species of heterogeneous resistant, intermediate and resistant bacteria in different parts of the world and it has created a great concern in clinical samples so that the number of the species of these vancomycin resistant bacteria are increasing.
Being vancomycin resistant, these bacteria are usually become resistant to a wide range of antibiotics. VISA (Vancomycin Intermediate resistance Staphylococcus aureus) and VRSA species have the potential for development and they are also increasing in our country. Vancomycin resistant staphylococcus aureus is at the head of research topics because the increasing use of surgeries, dialysis and modern medical methods lead to the increasing use of vancomycin. Nowadays there are few drugs like vancomycin which cover this bacterium. Due to the above mentioned reason, this research was done in this regard. It is obvious that studying multi-drug resistance like simultaneous resistance to vancomycin and tetracycline, pave the way to detect resistant species more effectively. It is accepted as a general rule that tetracycline resistance causes resistance to other antibiotics [7].
In this research, van (A) gene was not observed among 150 staphylococcus aureus samples. This result can be satisfying, for being vancomycin resistant to is really worrying and dangerous. At the moment, vancomycin is one of the few antibiotics which are effective in the treatment of staphylococcus aureus infections. The observed vancomycin resistance of this bacterium through disk diffusion method can be the result of phenotypic causes or environmental and human error. In addition the results can indicate that disk diffusion method is not accurate enough for this research. This result is consistent with the findings of some researchers in the field. Hong Binkim et al. (1999-2001) studied 682 staphylococcus aureus samples in Korea and no resistant or intermediate resistant sample was observed [8,9]. In a research which was done in Be sat hospital in Sanandaj, there was no vancomysin resistant strain or VRSA [10]. In another research, 5/6% was vancomycin resistant enterococcus but there was no VISA or VRSA [11]. In this research, 94/87% of vancomysin intermediate resistant staphylococcus aureus samples (74 out of 78) are resistant to methicillin (through disk diffusion). Also 94/44% of vancomycin resistant staphylococcus aureus samples (17 out of 18) are considered as MRSA samples through disk diffusion method, because most of vancomycin resistant, intermediate resistant are resistant to methicillin as well. Lack of resistant gene expression in all resistant samples to that gene, can be due to the different reasons, including phenotypic method or environmental and human error. In addition, observing phenotypic resistance is not a definite reason for resistant gene expression. In other words, lack of resistant gene in some antibiotic resistant samples could be also due to the lack of gene expression. For example, in studying van (A) gene, 12% of staphylococcus samples were identified as resistant ones through phenotypic method. In PCR stage, there was no resistant gene so it can be applicable to the above mentioned content. Huys et al. reported 58% of tet (K) gene in collected staphylococcus aureus from poultry slaughterhouses in South Africa [12]. In this research the prevalence of tetracycline resistance due to tet (K) gene was higher than other genes and this result is consistent with other researcher’s findings in this field. The lowest resistance to tetracycline was related to tet (O) gene which is consistent with other researchers’ findings too. As mentioned before, those staphylococcus aureus strains which only have tet (K) gene are tetracycline resistant, but they are susceptible to minocycline. Those strains which have tet (M) gene are resistant to all antibiotics in doxycycline group. These strains have tet (K) gene as well. Tet (L) gene is observed in the strains which have tet (M) gene. There was no report of tet (O) gene in staphylococcus aureus strains until 2004. Staphylococcus aureus has a wide distribution of tet (K) and tet (M) genes. These two genes have been located on mobile genetic elements such as small plasmids and transposons. It should be mentioned that this small plasmid is not able to transfer spontaneously but it can get the ability of movement from plasmids having conjugative genetic transmission power [13]. In addition, in this research 21 out of 32 tet (M) staphylococcus aureus samples (65/62%) have tet (K) gene as well. According to Greet Huys [12], staphylococcus aureus has a wide distribution of tet (K) and tet (M) genes which is in consistent with the result of this research. Also a tet (O) staphylococcus aureus strain has tet (K) as well. 3 out of 5 samples of tet (L) staphylococcus aureus have tet (K) too (60%). Only 1 out of 5 samples of tet (L) staphylococcus aureus, has tet (M) gene (20%). Among 150 staphylococcus aureus samples, only one sample had three tet (K), tet (M) and tet (L) genes (66% of all staphylococcus aureus samples). In other words, these three genes were expressed just in one sample.
Conclusion
This study indicates that resistance to tetracycline is mainly by efflux pumps mediated by tet (K) in Staphylococcus aureus in Sanandaj.
Author’s Contributions
Fatemeh keshavarzi designed the study, Fatemeh keshavarzi and Fatemeh Aeini wrote the paper; Foozieh Arabzadeh and Pezhman Karami, conceived the experiments, prepared the Figures; collected the samples. All authors gave final approval for the manuscript to be submitted for publication.
23536
References
- Franklin D, Lowy MD (1998) Staphylococcus aureus Infection. N Engl J Med 339: 520- 532.
- Hauschild T, Kehrenberg C, Schwarz CT (2003) Etracycline resistance in staphylococci from free-living rodents and insectivores. J Vet Med Series B Infect Dis Vet Publ Health 50: 443-446.
- Akoua Koffi C, Dje K, Toure R, Guessennd N, Acho B, et al. Nasal carriage of methicillin-resistant Staphylococcus aureus among health care personnel in Abidjan. Dakar Med 49(1): 70-74.
- Chopra I, Roberts M (2001) Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65(2): 232-260.
- Klein E, Smith DL, Laxinarayan R (2007) Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aurus, United states, Emerg Infect Dis 13(12): 1840-1860.
- Jones CH, Tuckman M, Howe AY, Orlowski M, Mullen S, et al. (2006) Diagnostic PCR analysis of the occurrence of methicillin and tetracycline resistance genes among Staphylococcus aureus isolates from phase 3 clinical trials of tigecycline for complicated skin and skin structure infections. Antimicrob Agents Chemother 50(2): 505-510.
- Locksley RM (2005) Staphylococal infections in Harrison's principles of internal medicine, 16th Edition, volum I, United States of America, Mc Graw-Hill companies 745-768.
- Kim HB, Park WB, Lee KD, Choi YJ, Park SW, et al. (2003) National Surveillance for Staphylococcus aureus with reduced susceptibility to Vancomycin in Korea. J Clin Microbiol 41(6): 2279-2281.
- Wang G, Hindler GF, Ward KW, Brunckner DA (2006) Increased vancomycin MICs for Staphylococcus aureus clinical isolates from a university hospital during a 5 year period. J Clin Microbiol 44(11): 3883-3886.
- Rashidian M, Taherpoor A, Goodarzi S (2001) Nasal carrier rates and antibiotic resistance of staphylococcus areus isolates of Beasat hospital staff. Scientific Journal of Kurdistan University of Medical Sciences 6(1): 1-8.
- Ghasemian R, Najafia N, Shojai A (2004) Nasal carriage and antibiotic resistance of staphylococcus areus isolates of Razi hospital personel, Qaemshahr. J Mazandaran Univ Med Sci 14(44): 79-86.
- Huys G, Cnockaert M, Vaneechoutte M, Woodford N, Nemec A, et al. (2005) Distribution of tetracycline resistance genes in genotypically related and unrelated multiresistant Acinetobacter baumannii strains from different European hospitals. Res Microbiol 156(3): 348-355.
- Tyagi S, Oberoi A (2015) Prevalence of inducible clindamycin resistance among Staphylococcal isolates in a tertiary care hospital in North India Indian J Med Microbiol 33: 327-328.