Keywords
Actinomycetes; Fibrinolytic enzyme; pH stability; Temperature stability
Introduction
Intravascular thrombosis is a major precipitating factor for cardiovascular diseases, accounting for about 17.3 million deaths annually, representing 30% of total mortality rate globally [1-3]. Currently used fibrinolytic (thrombolytic) agents e.g. tissue-type plasminogen activator (t-PA), urokinase and streptokinase convert plasminogen to plasmin that degrades the fibrin clot but suffer from shortcomings such as low specificity and stability, allergic reactions, resistance to repercussion, haemorrhagic side effects, large therapeutic doses and high costs, though some fibrinolytic agents can be direct acting by mimicking plasmin [4]. This warrants the search for novel fibrinolytic enzymes from various sources with higher efficacy, safety, specificity and stability and preferably those direct acting activities. Though new fibrinolytic enzymes are being explored from microbes [5], animals [6], plants [7] and fermented foods [8], microorganisms remain the preferred source due to their biochemical versatility, feasibility of mass culture and ease of genetic manipulation.
Actinomycetes, in spite of being excellent producers of bioactive and structurally novel metabolites, only a few fibrinolytic Streptomyces species have been reported in the literature e.g. Streptomyces sp. NRC 411 [9], Streptomyces megasporus SD5 [10] and Streptomyces sp. CS684 [11]. Most studies so far have been focused on Bacillus species isolated from both food and non-food sources e.g. Bacillus natto from Japanese fermented soybean food, natto [12], Bacillus sp. KA38 from Korean fermented fish, jeot-gal [13] and Bacillus amyloliquefaciens DC-4 from Chinese’s douchi [14]. Thus, the present work aims to screen and characterize fibrinolytic enzymes from microorganisms especially actinomycetes that are deposited in MBRL culture collection.
Materials and Methods
Strains and cultivation
The actinomycete strains for the present study were obtained from Microbial Biotechnology Research Laboratory (MBRL) culture collection, Manipur University, India and Lactic Acid Bacteria (LAB) strains from Microbial Biotechnology Laboratory, North Eastern Hill University, India. The cultures were revived on nutrient agar plate at 30°C for 7 days. A loopful of cultures were scraped from the plate and inoculated into seed medium [Glucose/Yeast extract/Peptone (GYP) broth] containing (% w/v): glucose, 1; yeast extract, 0.5; peptone, 0.5; NaCl, 0.5 and CaCl2, 0.02. They were grown for 5 days in shaker incubator (180 rpm) at 30°C. Cells were harvested by centrifugation at 10,000 rpm for 15 min at 4°C. The culture supernatants were filtered through sterile Whatman filter paper no.1 and the cell free filtrates were assayed for enzyme activity.
Screening of fibrinolytic activity
Screening of the enzyme activity was done by plasminogenfree fibrin plate assay [15]. The plate was made up of fibrinogen solution (2.5 ml of 1.2% fibrinogen) in 0.1 M sodium phosphate buffer, pH 7.4), 10 U thrombin and 1% agarose. 100 μl of the cell free filtrates was carefully placed on the plates and the zones of clearance were checked after incubation at 37°C for 18 hrs.
Fibrinolytic enzyme extraction
The actinomycete and LAB isolates were grown on GYP broth for 5 day at 30°C on a shaker incubator. Cells were harvested by centrifugation at 10,000 rpm for 15 min at 4°C. The cell free supernatants were subjected to ammonium sulphate (20-80% w/v) precipitation with constant overnight stirring at 4°C. The precipitate formed was centrifuged at 10,000 rpm for 30 min and the precipitate obtained was dissolved in 50 mM Tris-HCl buffer (pH 7.2). The dark brown solution thus obtained was dialyzed at 4°C for 24-48 hr against 10 mM Tris-HCl buffer, pH 7.2 [16]. The dialyzed crude was stored at 4°C for further analysis of fibrinolytic enzyme activity.
Assay of fibrinolytic activity
Fibrinolytic activity was estimated according to protocols followed by Raju and Divakar [17]. Briefly, 1.4 ml of 50 mM Tris–HCl (pH 8.0) and 0.4 ml of 0.72% (w/v) fibrinogen solution were mixed and incubated at 37°C for 5 min. Then 0.1 ml (20 U/ ml) of thrombin was added and the mixture was kept at 37°C for 10 min. Enzyme sample (0.1 ml) was added and incubation was done for an additional 60 min. The reaction was stopped by addition of 0.2 M trichloroacetic acid. After centrifugation (10,000 rpm for 5 min), the absorbance of the supernatant (Ar) and blank (Ac) was measured at 275 nm and recorded. One unit of fibrinolytic activity (FU) is defined as the amount of enzyme required to produce an increase in absorbance equal to 0.01 in 60 min at 275 nm. Fibrinolytic unit (FU) = [(Ar-Ac) × dilution ratio of sample] / [0.001 × 60 × 0.1].
Effect of temperature on enzyme activity and stability
The effect of temperature on enzyme activity was determined by incubating the enzyme in 50 mM sodium phosphate buffer, pH 7.0 for 5 hrs at various temperatures (-20 to 100°C). The thermal stability of each enzyme solution was determined by measuring the residual activities after incubating the enzyme for 60 min at 37°C [18].
Effect of pH on enzyme stability
The effect of pH on enzyme activity was determined by incubating the enzyme (at ratio of 1:1) at different pH ranging from 3.0–11.0 at 37°C for 5 hrs [19]. The buffer systems used were 0.2 M citrate buffer (pH 3.0 and 5.0), 0.2 M Tris-HCl buffer (pH 7.0) and 0.2 M glycine NaOH buffer (pH 9.0 and 11.0). pH stability was determined by measuring the residual activities after incubating the enzyme for 60 min at 37°C.
Results
Screening of fibrinolytic enzyme
A total of 33 strains, obtained from different habitats of North-East India, were screened for fibrinolytic activity of which 12 isolates showed positive results (Table 1 and Figure 1). Of the 12 fibrinolytic isolates, 5 strains (NRB1-19, MBRL-575 NRSI-19, SxF2, SxL6 and FS19) showed very potent activity with zone of clearance above 21 mm. SxF2 and SxL6 are endophytic actinomycetes, and FS19, NRBI-19 and MBRL-575 are LAB, actinomycete and Bacillus sp. respectively.
| Sl. No. |
Isolates |
Source |
Fibrinolytic enzyme activity |
| 1. |
NRP1-14 |
Nambul river |
- |
| 2. |
NRP1-26 |
Nambul river |
- |
| 3. |
NRP1-35 |
Nambul river |
- |
| 4. |
NRB1-19 |
Nambul river |
+ ++ |
| 5. |
NRB1-44 |
Nambul river |
- |
| 6. |
NRS1-18 |
Nambul river |
+ |
| 7. |
NRS1-116 |
Nambul river |
- |
| 8. |
LS1-81 |
Loktak lake |
- |
| 9. |
LS1-88 |
Loktak lake |
- |
| 10. |
LS1-128 |
Loktak lake |
- |
| 11. |
LS1-145 |
Loktak lake |
- |
| 12. |
LSCH-10C |
Loktak lake |
- |
| 13. |
LWG-2 |
Agricultural soil |
+ |
| 14. |
HA2 |
Hundung |
- |
| 15. |
HA4 |
Hundung |
+ |
| 16. |
MBRL575 |
Hundung limestone deposits |
+++ |
| 17. |
SJ-1 |
Shirui jungle |
- |
| 18. |
SJ-2 |
Shirui jungle |
- |
| 19. |
SJ-3 |
Shirui jungle |
- |
| 20. |
SL2 |
Shirui hills |
- |
| 21. |
SL3 |
Shirui hills |
- |
| 22. |
SK1-1 |
Salt springs |
- |
| 23. |
SK1-3 |
Salt springs |
- |
| 24. |
BLU-3 |
Bamboo leaves |
- |
| 25. |
BLU-5 |
Bamboo leaves |
- |
| 26. |
SxF1 |
Solanumxanthocarpum |
+++ |
| 27. |
SxF2 |
Solanumxanthocarpum |
+ |
| 28. |
SxL6 |
Solanumxanthocarpum |
++ |
| 29. |
SxL10 |
Solanumxanthocarpum |
+++ |
| 30. |
ANR |
Artemesianilagarica |
- |
| 31. |
FS1 |
Fermented soybean food |
++ |
| 32. |
FS7 |
Fermented soybean food |
++ |
| 33. |
FS19 |
Fermented soybean food |
+++ |
- : no activity + : activity with zone size between 10-15mm
++ : activity with zone size between 16-20mm
+++ : activity with zone size 21mm and above |
Table 1: Fibrinolytic activity profile.
Figure 1: Plasminogen-free fibrin plate assay for screening fibrinolytic activity, Zone of clearance indicates hydrolysis of fibrin clots.
Extraction and assay of fibrinolytic activity
The fibrinolytic enzymes were extracted by ammonium sulphate precipitation followed by dialysis. It was found that, for most of the isolates, the precipitation occurred at 50% (w/v) saturation except for SxF1, SxF2, SxL6 and SxL10 (endophytic actinomycetes) for which precipitation was achieved at 80% (w/v) saturation. The obtained dialyzed crude enzymes were used for the assay of fibrinolytic activities. MBRL-575 and NRBI-19 strains showed the highest activity with 33.66 U/ml and 33.48 U/ml respectively (Figure 2). FS7 and FS19 also showed high activity with 19.38 U/ml and 20.05 U/ml respectively. SxF1 which had very high activity on plate showed less activity with 8.1 U/ml when assayed.
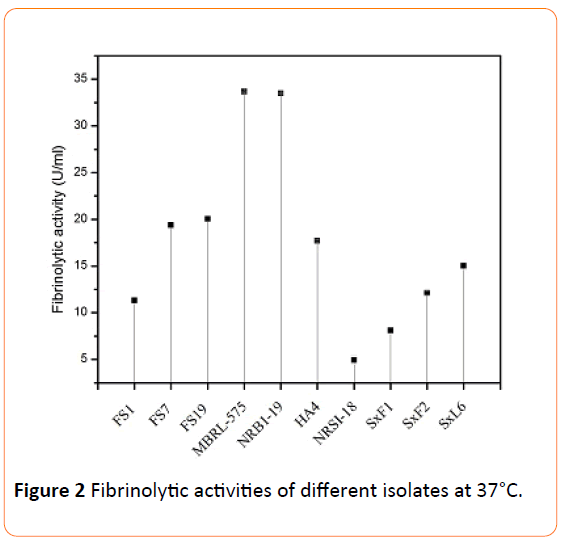
Figure 2: Fibrinolytic activities of different isolates at 37°C.
Effect of temperature
The activity and stability of the crude enzymes were determined at various temperatures between -20 and 100°C at pH 7 for 5 hrs. Most of the enzymes were active at temperature range of -20 to 55°C (Table 2 and Figure 3). The maximum activities were observed at 27 and 40°C, however as the temperature increases towards 55°C their activities gradually decreased and was completely lost at 100°C.
| Isolates |
Organism |
Source |
Optimal |
Stability |
Specific activity (U/ml) |
| Temperature (°C) |
pH |
Temperature (°C) |
pH |
| MBRL-575 |
Bacillus sp. |
Hundung limestone deposits |
40 |
9 |
-20 - 55 |
7 - 9 |
33.66 |
| NRB1-19 |
Actinomycetes |
Nambul river |
40 |
7 |
-20 - 55 |
7 - 9 |
33.48 |
| FS19 |
Bacillus sp. |
Fermented soybean food |
27 |
7 |
-20 - 40 |
7 - 9 |
20.05 |
| FS7 |
Vagococcus sp. |
Fermented soybean food |
27 |
7 |
-20 - 40 |
7 - 9 |
19.38 |
| HA4 |
Actinomycetes |
Hundung limestone deposits |
27 |
7 |
10 - 55 |
7 - 9 |
17.68 |
| SxL6 |
Actinomycetes |
Solanumxanthocarpum |
27 |
7 |
-20 - 40 |
5 - 9 |
15.02 |
| SxF2 |
Actinomycetes |
Solanumxanthocarpum |
27 |
7 |
-20 - 40 |
7 - 9 |
12.1 |
| FS1 |
Lactococcussp. |
Fermented soybean food |
27 |
7 |
-20 - 40 |
5 - 9 |
11.3 |
| SxF1 |
Actinomycetes |
Solanumxanthocarpum |
27 |
7 |
10 - 40 |
5 - 9 |
8.1 |
| NRSI-18 |
Actinomycetes |
Nambul river |
40 |
7 |
-20 - 40 |
5 - 9 |
4.93 |
Table 2: Biochemical profile of fibrinolytic enzymes obtained from different sources.

Figure 3: Effect of temperature on the fibrinolytic enzyme activity.
Effect of pH
To study the effect of pH on fibrinolytic activity and enzyme stability, assays were conducted at different pH ranging from 3.0-11.0 at 37°C for 5 hrs. The effects of pH on fibrinolytic activity and enzyme stability are shown in Figure 4 and Table 2. The enzymes were highly stable at pH 7 to 9. At pH 3 and 5 the activities were very less. However, the activity increased as the pH rises from 7 to 9, and then decreased at pH 11. The optimum pH of the enzyme was 7.0 except for MBRL-575 which was 9.
Discussion
In the past few decades, many fibrinolytic enzymes have been reported from various microbes including actinomycetes eg. Streptomyces sp. NRC 411 [9], Streptomyces sp. [20], Actinomycetes [21,22], Bacillus natto [12], Staphylococcus aureus [23], Bacillus sp. CK [24], Bacillus sp. KA38 [13] and Bacillus sp. strain DJ-4 [1]. Fibrinolytic enzymes have also been reported from different, such as fermented foods [8,12], soils [22], animals [6] and marine algae [7]. Endophytic strain Paenibacillus polymyxa EJS-3 has also been reported to secrete two extracellular fibrinolytic enzymes (118 and 49 kDa) in culture broth [25]. In the present study 1 LAB, 2 Bacillus sps. and 3 actinomycete strains were found to be potent producers of fibrinolytic enzymes. The specific activity of these isolates ranged from 15.02 to 33.66 U/ml. Simkhada et al. [11] reported a novel fibrinolytic protease from Streptomyces sp. CS684 that had a specific activity of 19 U/mg. Ju et al. [22] purified a fibrinolytic enzyme from Streptomyces sp. XZNUM 00004 that has a specific activity of 530.0 IU/mg. Hyeon-Deok et al. [26] and Chang et al. [27] reported fibrinolytic enzyme from Bacillus subtilis and Bacillus amyloliquefaciens MJ5-41 that had a specific activity of 21.6 U/mg and 3.44 U/mg respectively.
The temperature stability profile of the enzymes ranged from -20 to 55°C. It was found that enzymes of MBRL-575, FS7, FS19 and SxL6 could retain more than 50% activity at 40°C even after 5 hrs of incubation. NRB1-19, however, could retain only 33% of its initial activity. At temperature 55°C the enzymes retained only about 23% activity. Mahajan et al. [5] reported that the purified enzyme of Bacillus subtilis ICTF-1, after 1 hr of incubation at 40°C and 50°C, retained 50% and 18% of its initial activity respectively. Agrebi et al. [28] reported a fibrinolytic enzyme from Bacillus amyloliquefaciens which had relatively higher thermal stability. This enzyme retained 62% of its initial activity at 50°C after 1 hr incubation but was completely inactivated after 20 min at 60°C and 2 min at 70°C. Chitte and Dey [10] extracted a fibrinolytic enzyme from a thermophilic Streptomyces megasporus strain SD5 that had more than 66% activity at 37°C.
The enzyme stability at different pH for the present study ranged from pH 5-9. The optimal pH was found to be 7.0 except for MBRL-575 which was 9. It was found that MBRL-575, which had the highest activity, could retain 50% of its initial activity at pH 9 after incubation for 5 hrs. NRB1-19, on the other hand, could retain only 21% of its initial activity. Many of the fibrinolytic enzymes reported are found to be stable at pH ranging from 4 -12, for instance Hassanein et al. [29] reported a fibrinolytic enzyme from Bacillus subtilis K42 that has maximum activity at pH 9.4 (stable at pH 6.5-10.5). Mander et al. [30] reported a fibrinolytic enzyme from Streptomyces sp. CS624 whose optimum pH was 7. This enzyme retained <90% activity at pH 6.5 and 7.5 after incubation at 4°C for 24 hrs. Mahajan et al. [5] reported an enzyme from Bacillus subtilis ICTF-1 that was stable at pH range of 7.0-11.0 but exhibited maximum activity at pH 9.0. The enzyme was also stable at pH range of 5.0-11.0 after incubation at 30°C for 60 min. Ko et al. [31] and Wang et al. [32] also reported fibrinolytic enzyme from Bacillus subtilis that has optimal activity at pH 8.
From the present study, it can be concluded that the fibrinolytic enzymes, especially MBRL-575, can withstand alkaline pH and cold temperature, and can be preserved at -20°C. The enzymes are stable at pH range of pH 5-9 and temperature ranging from -20 to 55°C.
Acknowledgments
The authors are thankful to Dr. SR Joshi, North-Eastern Hill University, Shillong, Meghalaya, for providing some of the cultures used in the present study.
Funding
This study was supported by the Department of Biotechnology (DBT), Ministry of Science & Technology, and Government of India by providing DBT RA fellowship to ST. We also gratefully acknowledge the grant from the Department of Biotechnology (DBT), Government of India, under the State Biotech Hub (SBT Hub) Scheme (BT/04/NE/2009) for the material and infrastructural support that facilitated part of this research work.
Conflicting Interests
The authors declare that they have no conflicting interests.
9419
References
- Kim SH, Choi NS (2000) Purification and characterization of subtilisin DJ-4 secreted by Bacillus sp. strain DJ-4 screened from Doen-jang. BiosciBiotechnolBiochem 64: 1725-1772.
- Mine Y, Won AHK, Jiang B (2005) Fibrinolytic enzymes in Asian traditional fermented foods. Food Res Int 38: 243-250.
- WHO (2008) https://www.who.int/gho/publications/world_health_statistics/EN_WHS08_ Full.pdf.
- Nakajima N, Mihara H, Sumi H (1993) Characterization of potent fibrinolytic enzymes in earthworm, Lumbricusrubellus. BiosciBiotechnolBiochem 57: 1726-1730.
- Mahajan PM, Nayak S, Lele SS (2012) Fibrinolytic enzyme from newly isolated marine bacterium Bacillus subtilis ICTF-1: Media optimization, purification and characterization. J BiosciBioeng 113: 307-314.
- Wang D, Liu W, Han B, Xu R (2007) Biochemical and enzymatic properties of a novel marine fibrinolytic enzyme from Urechisunicinctus. ApplBiochemBiotechnol 136: 251-264.
- Matsubara K, Hori K, Matsuura Y, Miyazaw K (1999) A fbrinolytic enzyme from a marine green alga Codiumlatum. Phytochem 52: 993-999.
- Jaeyoung P, Yoon S, Kim S, Lee B, Cheong H (2012) Characterization and fibrinolytic activity of Acetobacter sp. FP1 isolated from fermented pine needle extract. J MicrobiolBiotechnol 22: 215-219.
- Abdel-Naby MA, El-Diwany AL, Shaker HM, Ismail AMS (1992) Production and properties of a fibrinolytic enzyme from Streptomyces sp. NRC 411. MicrobiolBiotechnol 8: 267-269.
- Chitte RR, Dey S (2000) Potent fibrinolytic enzyme from a thermophilic Streptomyces megasporus strain SD5.LettApplMicrobiol 31: 405-410.
- Simkhada JR, Mander P, Cho SS, Yoo JC (2010) A novel fibrinolytic protease from Streptomyces sp. CS684. ProcBiochem 45: 88-93.
- Sumi H, Hamada H, Tsushima H, Mihara H, Muraki H (1987) A novel fibrinolytic enzyme (nattokinase) in the vegetable cheese Natto; a typical and popular soybean food in the Japanese diet. Experientia 43: 1110-1111.
- Kim HK, Kim GT, Kim DK, Choi WA, Park SH, et al. (1997) Purification and characterization of a novel fibrinolytic enzyme from Bacillus sp. KA38 originated from fermented fish. J Ferment Bioeng 84: 307-312.
- Peng Y, Huang Q, Zhang RH, Zang YZ (2003) Purification and characterization of a fibrinolytic enzyme produced by Bacillus amyloliquwfaceiens DC-4 screened from a traditional Chinese soybean food. Comp BiochemPhysiolBiochemMolBiol 134: 45-52.
- Jeong YK, Yang WS, Kim KH, Chung KT, Joo WH, et al. (2004) Purification of a fibrinolytic enzyme (myulchikinase) from pickledanchovy and its cytotoxicity to the tumor cell lines. BiotechnolLett 26: 393-397.
- Balaraman K, Prabakaran G (2007) Production and purification of a fibrinolytic enzyme (thrombinase) from Bacillus sphaericus. Ind J Med Res 126: 459-464.
- Raju EVN, Divakar G (2013) Bacillus Cereus GD 55 strain improvement by physical and chemical mutagenesis for enhanced production of fibrinolytic protease. Int J PharmaSci Res 4: 81-93.
- Cha WS, Park SS, Kim SJ, Choi DB (2010) Biochemical and enzymatic properties of a fibrinolytic enzyme from Pleurotuseryngii cultivated under solid-state conditions using corn cob. BioresTechnol 101: 6475-6481.
- Pandee P, H-Kittikul A, Masahiro O, Dissara Y (2008) Production and properties of a fibrinolytic enzyme by Schizophyllum commune BL23. Songklanakarin J SciTechnol 30: 447-453.
- Chitte RR, Deshmukh SV, Kanekar PP (2011) Production, purification, and biochemical characterization of a fibrinolytic enzyme from thermophilic Streptomyces sp. MCMB-379. ApplBiochemBiotechnol 165: 1406-1413.
- Balachandran C, Duraipandiyan V, Ignacimuthu S (2012) Purification and characterization of protease enzyme from actinomycetes and its cytotoxic effect on cancer cell line (A549). Asian Pac J Trop Biomed 2: S392-S400.
- Ju X, Cao X, Sun Y, Wang Z, Cao C, Liu J, Jiang J (2012) Purification and characterization of a fibrinolytic enzyme from Streptomyces sp. XZNUM 00004. World J MicrobiolBiotechnol 28: 2479-2486.
- Lijnen HR, van Hoef S, de Cock F, Okada K, Ueshima S, et al. (1991) On the mechanism of fibrin-specific plasminogen activation by staphylokinase. J BiolChem 239: 11826-11832.
- Kim W, Choi K, Kim Y (1996) Purification and characterization of a fibrinolytic enzyme produced from Bacillus sp. strain CK 11-4 screened from Chungkook- Jang. ApplEnvMicrobiol 62: 2482-2488.
- Lu F, Sun L, Lu Z, Bie X, Fang Y, Liu S (2007) Isolation and identification of an endophytic strain EJS-3 producing novel fibrinolytic enzymes. CurrMicrobiol 54: 435-439.
- Hyeon-Deok J, Lee HA, Jeong SJ, Kim JHJ (2011) Purification and characterization of a major fibrinolytic enzyme from Bacillus amyloliquefaciens MJ5-41 isolated from Meju. MicrobiolBiotechnol 21: 1166-1173.
- Chang CT, Wang PM, Hung YF, Chung YC (2012) Purification and biochemical properties of a fibrinolytic enzyme from Bacillus subtilis-fermented red bean. Food Chem 133: 1611-1617.
- Agrebi R, Hmidet N, Hajji M, Ktari N, Haddar A, et al. (2010) Fibrinolytic serine protease isolation from Bacillus amyloliquefaciens An6 grown on Mirabilis jalapa tuber powders. ApplBiochemBiotechnol 162: 75-88.
- Hassanein WA, Kotb E, Awny NM, El-Zawahry YA (2011) Fibrinolysis and anticoagulant potential of a metallo protease produced by Bacillus subtilis K42. J Biosci 36: 1-7.
- Mander P, Cho SS, Simkhada JR, Choi YH, Yoo JC (2011) A low molecular weight chymotrypsin-like novel fibrinolytic enzyme from Streptomyces sp. CS624. ProcBiochem 46: 1449-1455
- Ko JH, Yan JP, Zhu L, Qi YP (2004) Identification of two novel fibrinolytic enzymes from Bacillus subtilis QK02. Comp BiochemPhysiol C ToxicolPharmacol 137: 65-74.
- Wang C, Ji B, Li B, Ji H (2006) Enzymatic properties and identification of a fibrinolytic serine protease purified from Bacillus subtilis DC33. World J MicrobiolBiotechnol 22: 1365-1371.








