Introduction
Lactobacilli are Gram-positive, mostly facultative but under certain conditions strictly anaerobic, non-sporulating rods. These bacteria have a long history of safe use, especially in the dairy industry [1]. Lactobacilli resemble a major part of the commensal human mucosal flora [2-6].Clinical studies could demonstrate a protective role of lactobacilli for intestinal infections and urogenital as well as a capability to prevent and treat allergic diseases [7,8]. Increased drive has existed for the isolation of novel Lactobacillus strains that exert a beneficial health effect when ingested by humans. Such strains are termed probiotic. Probiotics are defined as live microorganisms which when administered in adequate amounts confer a health benefit on the host [9].
In order for a probiotic strain to exert its beneficial effect on the host, it has to be able to survive passage through the host’s digestive tract [10]. The ability of Lactobacillus strains to adhere to the mucosal surfaces of the intestine and the subsequent colonization has long been one of the most commonly encountered criteria for the selection of probiotic strains. Adhesive probiotic lactobacilli have been reported to have beneficial health effects, especially related to the inhibition of pathogen adhesion to intestinal cell lines.
The aim of this study was to evaluate the probiotic potential of Lactobacillus isolates recovered from dairy products as well as infant stool by applying in vitro screening tests and to select probiotic candidates that fulfill the established criteria and could therefore be potentially used as novel therapeutic agents.
Material and Methods
Materials and their sources
Dolbeco modified Eagles Media (DMEM), Trypsin 0.25% - EDTA solution (sterile filtered), Fetal bovine serum (sterile filtered) was purchased from Sigma Aldrich Co.,USA. Rogosa agar (Oxide), deMan Rogosa Sharpe (MRS) broth (Difco), Bile Salts Mixture (Starchemic, India), Flat bottom sterile tissue culture plates of 96 wells (Nunclon, Denmark), BD BBL Gas- Pak™ 100 System, (Becton and Dickison company, USA),Gas Generating Kit (Oxoid).
Isolation and identification of Lactobacillus
Lactobacillus isolates were recovered from dairy products (raw milk, butter, cheese and yoghurt), stool specimens (from breast-fed infants) as well as freeze dried lactic culture (REDI-SET, used for Bulk starter culture, purchased from Chr. Hansen`s Lab., Denmark). Milk and processed milk products were obtained from farmers.
Isolation of Lactobacillus from dairy products
About 1 g of each sample (raw milk, cheese, yoghurt and butter) was taken aseptically, transferred to 10 ml phosphate buffered saline and then vortexed [11]. An aliquot (1 ml) of the homogenate produced was then inoculated in 10 ml MRS broth contained in screw caped tube, and incubated under anaerobic conditions (GasPak Anaerobic System) for 24 h at 37°C. An aliquot (50 μl) of the obtained culture was spread on acidified MRS agar plates, acidified with glacial acetic acid to pH 5.7 and incubated anaerobically for 48 h at 37°C. Colonies with typical characteristics of Lactobacillus (small, white, circular, entire margin with diameter 1-3 mm) were selected and subcultured in MRS broth for further purification and identification.
Isolation of Lactobacillus from infant stool
Lactobacilli were isolated from stools of healthy breast-fed infants (aged 3-6 months). One gram from a fresh fecal sample (maximum lag time was 2 h) was suspended in 9 ml of a 0.85% NaCl solution (pH 7.0). The slurry was homogenized and filtered to remove any large particles and debris [12]. For enrichment of lactobacilli, all the samples were diluted and cultured in deMan Rogosa Sharpe (MRS) broth at 37 °C for 48 h under anaerobic conditions, followed by plating on MRS or Rogosa agar. Isolated colonies with typical characteristics of lactobacilli were picked from the plates and further purified on MRS or Rogosa agar [13].
Recovery of Lactobacillus from Bulk starter culture
One gram of the dried lactic culture were suspended in 100 ml MRS broth, incubated at 37 °C for 48 h under anaerobic conditions, followed by subsequent subculture in MRS broth then, the obtained growth plated on acidified MRS agar plates (acidified with glacial acetic acid to pH 5.2). Isolated colonies with typical characteristics of lactobacilli were picked from the plates and further purified on MRS.
The collected isolates were stored at 20ºC in MRS broth containing 20% glycerol [14].
Identification of the selected colonies
The collected isolates were identified to genus level by microscopical examination and catalase and oxidase test [11,15-16].
Mammalian cell line
The cell line used in this study was Vero Cell Line (ATCC No. CCL-81), are kidney epithelial cells derived from African green monkey and was purchased from VACSERA, Egypt.
Screening the Lactobacillus isolates for their adherence capabilities to mammalian cells
Lactobacillus isolates were cultured anaerobically in MRS broth at 37°C. The 24-hours MRS culture of tested Lactobacillus was centrifuged, washed twice with phosphate buffered saline pH 7 and then resuspended in DMEM and the count was standardized to 108 CFU/ml in the same medium. To evaluate binding of Lactobacillus isolates to epithelial cells, Vero cells which supplied as a confluent monolayer in 96-well tissue culture plates was washed and then, 200 μl aliquots of 108 CFU /ml Lactobacillus in DMEM were added to each well. After 2 hours of incubation at 37°C, cell culture medium was aspirated off and cells were washed three times with DMEMPBS (1:1), (pH 7.4, 37°C) to remove non-bound bacteria. Cells were released from polystyrene wells by adding 0.125 ml aliquots of 0.05% trypsin-EDTA to each well and incubat ing the plate at 37°C for 30 minutes [17]. Serial dilutions of bacteria were plated on MRS agar and the agar plates were incubated anaerobically for 48 h at 37°C for subsequent CFU quantification.
Testing the survival of Lactobacillus isolates under conditions simulating the human GI tract
The tested Lactobacillus isolate was cultured anaerobically in MRS broth at 37°C. Bacterial cells from overnight (18 h) culture were harvested at 5000 rpm, 10 min, and then they were washed twice with PBS buffer, pH 7.2, before being suspended in the same buffer. The bacterial count was adjusted to 108 CFU/ml (optical density 0.2 at 600 nm).
Acid Tolerance
The resistance of the examined lactobacilli to low pH environment was tested as described by Maragkoudakis [18]. Aliquots of 100 μl of the prepared bacterial suspension were added to 10 ml quantities of MRS broth contained in screw caped tubes adjusted to pH 2 or 3 to mimic gastric acidity and pH 6.2 as control. Initial populations was about 106 CFU/ ml. Resistance to acid was assessed in terms of viable colony counts and enumerated after incubation at 37°C anaerobically for 1 and 3 h, reflecting the time spent by food in the stomach. An aliquot of 100 μl was taken at 0, 1 and 3 h from each tube of specified pH, serially diluted and then plated on MRS agar and incubated anaerobically for 48 h at 37°C.
Bile Tolerance
Tolerance of Lactobacilli to bile was evaluated by examining the survival of the tested isolates in presence of 0.3% w/v bile salt mixture (Starchemic, India) which mimics the bile concentration of the human gastrointestinal tract. Aliquots of 100 μl quantity of the prepared bacterial suspension were added to 10 ml quantities of MRS broth contained in screw caped tubes, pH 6.2, enriched with 0.3% w/v bile salts and MRS broth having pH 6.2 as control. Initial populations was about 106 CFU/ml. Resistance to bile was assessed in terms of viable colony counts and enumerated after incubation at 37 °C anaerobically for 1 and 4 h, reflecting the time spent by food in the small intestine. An aliquot of 100 μl was taken at 0, 1 and 4 h from each tube, serially diluted and then plated on MRS agar and incubated anaerobically for 48 h at 37°C [18]. All screening experiments were done in triplicates, the presented data were average of three values.
Results
Isolation and identification of Lactobacillus
From the different samples collected from various sources (raw milk, cheese, butter, yoghurt as well as infant stool), 89 bacterial isolates and 12 yeast isolates were recovered.
The isolates that showed the Lactobacillus characteristics according to colony morphology on MRS agar (appeared as small, white, circular, entire margin with diameter 1-3 mm), Gram staining (Gram positive bacilli, non-endospore forming), catalase negative, oxidase negative were selected. These isolates (52) included; 35 isolates were recovered from raw milk, 8 isolates were recovered from cheese, 7 isolates were recovered from butter, 1 isolate (LS) from a freeze dried lactic culture and 1 isolate (S1) was recovered from infant stool.
Adherence of different Lactobacillus isolates to Vero cells
The adherence of the collected Lactobacillus isolates (52 isolates) to Vero cells was investigated and the adhered cells were expressed as percentage of initial count. The adherence capabilities to Vero cells varied greatly among the tested Lactobacillus isolates, ranging from 0.001% to 6.4% (Figure 1).
The number of bacteria adherent per Vero cell (adherence capacity) of the tested Lactobacillus isolates was shown in Table 1. The tested Lactobacillus isolates were categorized according to their relative adherence capacities. About 50% of the tested isolates Lactobacillus showed good adherence, about 40% showed moderate adherence while, 10% having no adherence capacity to Vero cells.
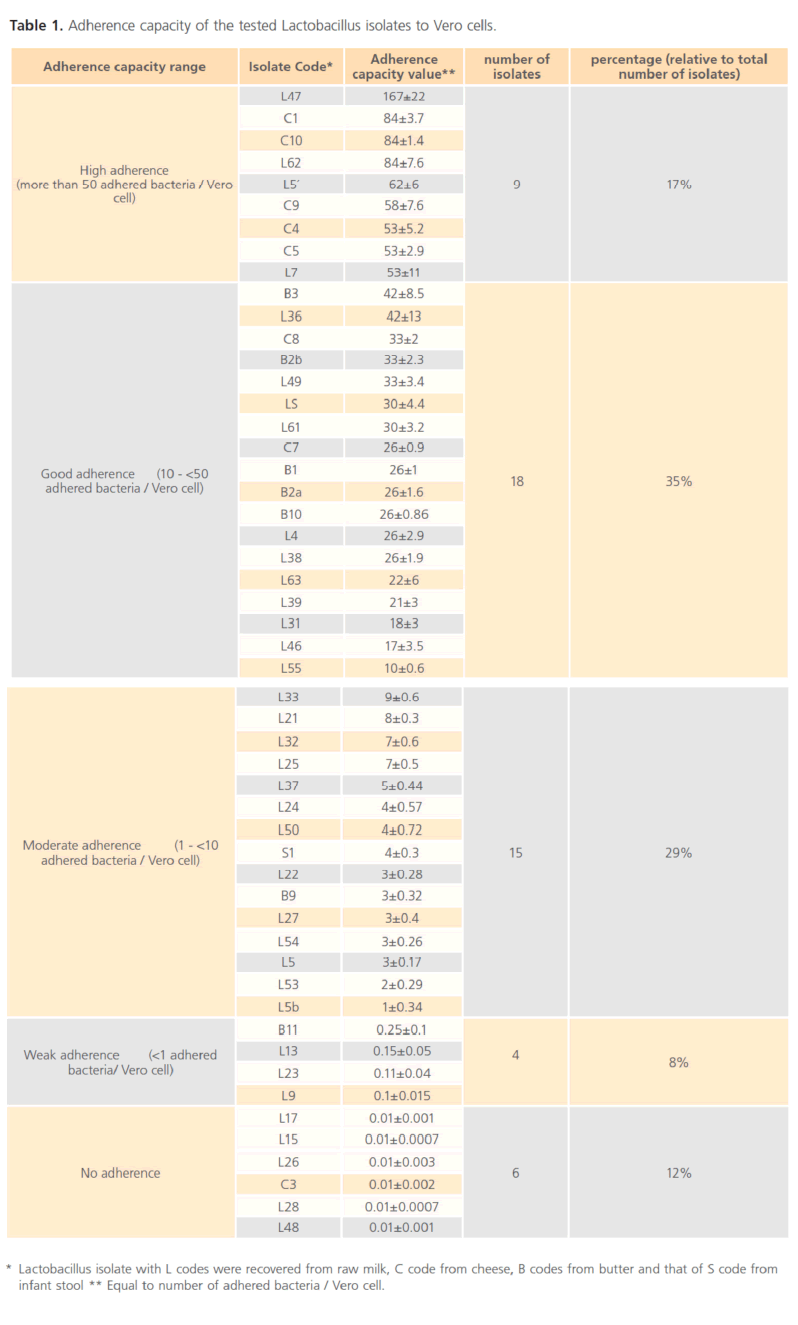
Table 1. Adherence capacity of the tested Lactobacillus isolates to Vero cells.
The selection of the Lactobacillus isolates for further testing was dependant on their adherence capacities to Vero cells. Isolates showed no adherence capacities to Vero cells were excluded, while the other isolates were further tested to investigate their probiotic potential.
Survival of the tested Lactobacillus isolates under conditions simulating GIT
The tolerance of Lactobacillus isolates, which showed adherence capacity > 0.1 adhered bacteria / Vero cell (46 isolates), to acid and bile was tested.
The resistance of these Lactobacillus isolates to acid was tested by examining their survival in MRS (pH 3) at 37°C after 0, 1, 3 h and compared with control (MRS pH 6.2). According to the obtained results (Figure 2), 44 out of 46 Lactobacillus isolates were able to survive after exposure to pH 3 for 3 h. Furthermore, 28 isolates (C1, C4, C7, C8, C9, C10, B1, B2a, B3, B10, B11, L4, L5’, L21, L24, L25, L27, L31,L39, L38, L37, L36, L33, L47, L49, L50, L53 & L55) were able to grow in MRS (PH 3) after 24 h incubation at 37°C giving turbidity comparable to that of the control when examined visually.
The survival of the 46 Lactobacillus isolates in MRS containing 0.3% bile salts (pH 6.2) at 37°C were examined at different time intervals (0, 1, 4 h) and compared with control (MRS pH 6.2). According to the obtained results (Figure 3), out of 46 Lactobacillus isolates, 33 isolates retained their viability after exposure to 0.3% bile salts for 4 h (reflecting the time spent in the small intestine). Furthermore, 15 isolates (C4, C7, C8, C9, C10, B2a, B2b, B3, B10, B11, L36, L38, L39, L47 & L49) were able to grow in the presence of 0.3% bile salts after 24 h incubation at 37°C giving turbidity comparable to that of the control when examined visually.
After screening of all isolates, we obtained 32 Lactobacillus isolates tolerant to stomach acidity and intestinal bile and having adherence capabilities to mammalian cells as shown in Table (2).
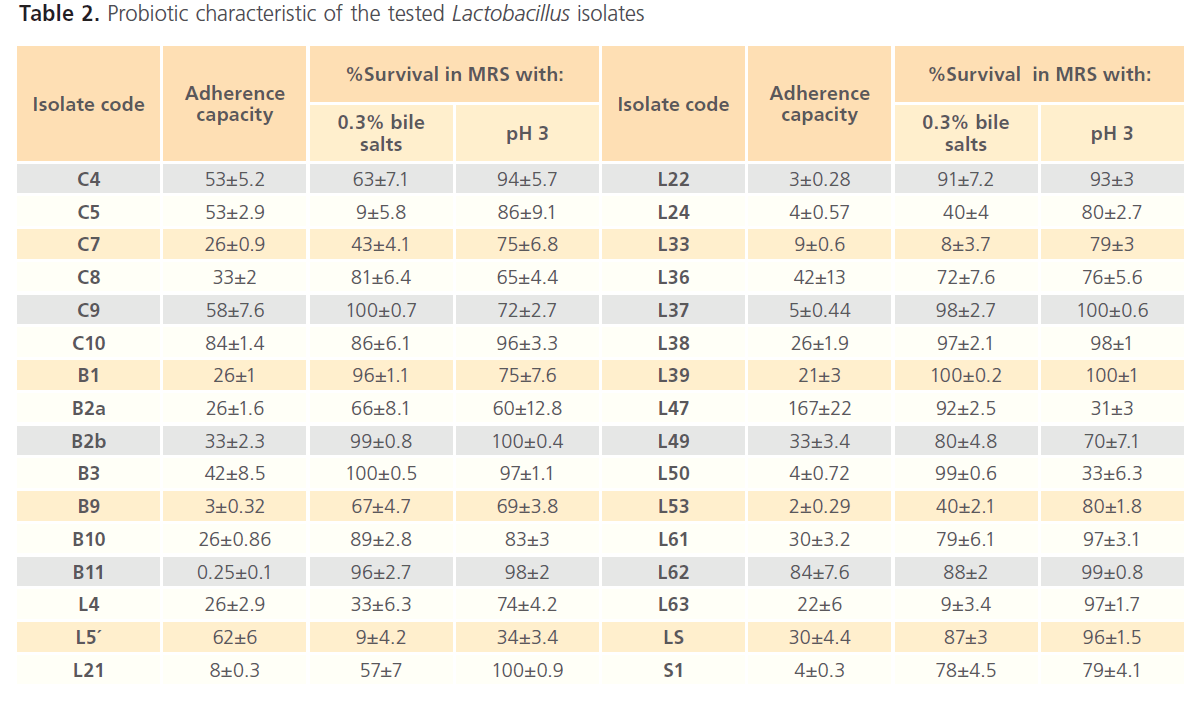
Table 2. Probiotic characteristic of the tested Lactobacillus isolates
Discussion
One of the most important criteria that a probiotic candidate should fulfill is its ability to adhere to the epithelial cells [15,19-20]. This property is important for colonization [21], pathogen exclusion, modulation of the immune system [22] and interaction with epithelial cells [23-24].
A variety of in vitro model systems for routine adhesion experiments e.g. Vero cell [25-26], Caco-2 & HT-29 [27] were used. In the present study, Vero cell was used as a model for investigation of bacterial adhesion to the normal epithelial cells (not cancerous cells). The adherence capacities of the tested Lactobacillus isolates (52 isolates) to Vero cells varied greatly(Figure 1 and Table 1), where nine Lactobacillus isolates (C1, C4, C5, C9, C10, L5’, L7, L47and L62) showed high adherence (more than 50 adhered bacteria per Vero cell), eighteen Lactobacillus isolates (C7, C8, B1, B2a, B2b, B3, B10, L4, L31, L36, L38, L39, L49, LS, L61, L63, L 46 and L55) showed good adherence (10 to 50 adhered bacteria per Vero cell), fifteen Lactobacillus isolates (L33, L21, L32, L25, L37, L24, L50, S1, L22, B9, L27, L54, L5, L53 and L5b) showed moderate adherence ( >1 adhered bacteria per Vero cell), four Lactobacillus isolates (B11, L13, L23 and L9) showed weak adherence ( <1 adhered bacteria per Vero cell) and six Lactobacillus isolates (L17, L15, L26, C3, L28 and L48) showed very poor adherence to Vero cells. Many studies showed that adhesive properties are not a universal feature of Lactobacillus because it varies considerably between Lactobacillus strains [21,28].
It is worth noting that some Lactobacillus isolates recovered from either raw milk or dairy products exhibited adherence to Vero cell higher than a representative intestinal Lactobacillus isolate (S1).
Several studies showed that lactobacilli have good capabilities to bind to different types of epithelial cells such as human vaginal epithelial cells, to cultured human carcinomal intestinal cell lines, to intestinal mucus, and to the components of the extracellular matrix (ECM) [28-41].
The adhesiveness of lactobacilli is strain-specific [8]. Several studies showed that the microbial adhesion process of lactobacilli includes passive forces, electrostatic interactions, hydrophobic steric forces, lipoteichoic acids; and specific structures [2,8,31,42-45]. McGroarty identified by transmission electron microscopy fimbriae on vaginal strains of L. rhamnosus, L. acidophilus, L. jensenii, L. casei, and L. fermentum. Fimbriated lactobacilli, in vitro, adhered in significantly greater numbers to human vaginal epithelial cells than those nonfimbriated variants [46]. Chan et al. suggested that lipoteichoic acid participates in the adherence of lactobacilli to uroepithelial cells [29], whereas Reid et al. identified two adhesins, an extracellular, probably proteinaceous, and a trypsininsensitive cell wall adhesion [47]. Boris et al. showed that three strains of lactobacilli, L. acidophilus, L. gasseri and L. jensenii strongly adhered to vaginal epithelial cells in vitro [31].
However, several ways of adherence exist: in L. acidophilus and L. gasseri proteins and carbohydrates participate in the adherence, whereas L. jensenii adherence seems to depend on carbohydrates alone. Sillanpaa and his workers reported that the S-layer protein from L. crispatus strains is involved in their adhesion [48]. For strains of L. plantarum, isolated from the human gastrointestinal tract, an expression of a mannose binding adhesin could be shown [2]. These findings confirm the presumption that different strains of lactobacilli differ in their capacity and way of adherence.
After oral ingestion of probiotic bacteria, they encounter a number of human defense systems that are associated with secretions. These include gastric acid inducing a low pH in the stomach, and bile salts secreted into the luminal content in the proximal small intestine [49].
Adherence to epithelial cells, resistance to gastric acidity and resistance to bile salts are among the in vitro tests that are frequently suggested for the evaluation of the probiotic potential of a bacterial strain [50-51]. In the present study, the 6 Lactobacillus isolates with no adherence capacity were excluded and the remaining 46 isolates were selected to investigate their probiotic potential, which included their acid and bile tolerance and antagonistic activity against some human pathogens. The tolerance to stomach acidity was investigated by testing the survival of Lactobacillus isolates in MRS at pH 3 which mimic the stomach pH. The survival also examined at different time intervals which reflect the time spent by food in the stomach. The results showed that only two isolates out of 46 can’t survive in acidic pH after exposure for 3 hours, while the remaining isolates showed acid tolerance expressed as survival percentage in acidic medium pH3 ranged between 30 – 100% survival (Table 2). These survival capabilities could be categorized into high survival percentage (75 – 100%), good survival percentage (50 - <75%), and moderate survival percentage (25 - <50%).
It is worth noting that the lowest survival percentage (21%) of the acid tolerant 44 Lactobacillus isolates didn’t exceed one Log10 reduction of initial count. Furthermore, prolonged incubation in the acidic medium of pH3 was overcome by about 64% of the acid tolerant isolates and resulted in an increase in growth after 24h.
These results were in agreement with those obtained from previous similar in vitro studies, where Lactobacillus strains were able to retain their viability when exposed to pH values of 2–3 [10,50,52-56]. However, Schillinger et al. found that Lactobacillus strains isolated from various probiotic dairy products differed considerably in their resistance to acid in a simulated gastric juice [27].
Conway et al. and Ouwehand et al. reported that survival of ingested bacteria in the stomach obviously is influenced by the buffering capacity of food components [10,57]. In this context, even strains not able to survive at pH2 in vitro could exhibit substantial viability when consumed in a matrix of fermented milk, where milk substances may provide a protective matrix enhancing survival of bacteria in the gastric juice.
Resistance to bile salts is generally considered as an important criterion for probiotic selection, since it is an essential property for probiotic strains to survive the conditions in the small intestine. The tolerance to bile was investigated by testing the survival of 46 Lactobacillus isolates in MRS enriched with 0.3% bile salts which mimic the physiological bile concentration [18]. The survival also examined at different time intervals which reflect the time spent by food in the small intestine. The results showed that 33 (about 70%) Lactobacillus isolates were able to survive in presence of bile after 4 hours exposure and could be considered as bile tolerant. These bile tolerant isolates showed different survival capabilities in MRS containing 0.3% bile salts after 4 h (Table 3) ranged between (8 – 100%). It could be relatively categorized into high survival percentage (75 – 100%), good survival percentage (50 - <75%), moderate survival percentage (25 - <50%) and weak survival percentage (20 - <50%).It is worth noting that and as observed for acid tolerance, 15 Lactobacillus isolates were able to grow in the presence of 0.3% bile after 24 h incubation at 37°C. In a similar study it was found that different strains of L. casei were able to grow in the presence of 0.5% bile [58]. Also, it was observed that twelve Lactobacillus isolates can’t survive in presence of 0.3% bile after 4 h, in spite of their acid tolerance (Tables 2 & 3).
Regarding acid and bile tolerance, the mechanisms involved in acid resistant of lactobacilli are not fully understood, but several studies detected some genes that are switched on under conditions simulating GIT. Bron and co-workers showed up regulation of stress proteins, cell envelope located proteins, and proteins involved in redox reactions after exposure of Lactobacillus strains to 0.1% porcine bile [59]. Many authors suggested that the resistance of Lactobacillus strains to the toxicity of bile salts in the duodenum may be attributed to bile salt hydrolytic (BSH) activities [60]. Conversely, other studies showed that resistance to bile salts and bile salt hydrolase activity are unrelated in lactobacilli [61]. Several in vitro studies showed alterations in the cell wall, presumably to protect the cell from the harsh conditions (acid and bile).
Interestingly and as observed in case of adherence, the results showed that the survival percentage of some Lactobacillus isolates recovered from dairy products after exposure to 0.3% bile salts was higher than that of Lactobacillus isolate S1 which is of intestinal origin. According to these findings, Lactobacillus isolates recovered from dairy products could be able to colonize and survive the harsh conditions encountered during the passage through GIT efficiently comparable or even superior to intestinal lactobacilli.
In agreement with the present study, different strains of L. plantarum were found to show a high tolerance to the consecutive exposure to hydrochloric acid (pH 2.0) and bile salts. This was observed both for strains isolated from intestinal samples and for those isolated from fermented foods [55].
To conclude, in this study thirty two Lactobacillus isolates were found to possess desirable probiotic properties. These isolates are good candidates for further investigation in in vitro and in vivo studies for their potential health benefits and their application as novel Biotherapeutic agents.
137
References
- Borriello SP, Hammes WP, Holzapfel W, Marteau P, Schrezenmeir J, “et al.” (2003) Safety of Probiotics That Contain lactobacilli or Bifidobacteria.Clin Infect Dis. 36:775–80
- Ahrne´ S, Nobaek S, Jeppsson B, Adlerberth I, Wold AE, Molin G (1998). The normal Lactobacillus flora of healthy human rectal and oral mucosa. J. Appl. Microbiol. 85: 88–94.
- Andreu A, Stapleton AE, Fennell CL, Hillier SL, Stamm WE (1995) Hemagglutination, adherence, and surface properties of vaginal Lactobacillus species. J. Infect. Dis.. 171, 1237–1243.
- Georgi A, Torriani S, Dellaglio F, Bo G, Stola E, Bernuzzi L (1987) Identification of vaginal lactobacilli from asymptomatic women. Microbiologica. 10:377–384.
- Holzapfel WH, Haberer P, Snel J, Schillinger U, and Huisin’t Veld JH (1998) Overview of gut flora and probiotics. Int. J. Food Microbiol. 41:85-101
- Tannock GW (1999) Analysis of the intestinal microflora: a renaissance. Antonie van Leeuwenhok. 76:265-278
- Merk K, Borelli C, Korting HC (2005). Lactobacilli – bacteria–host interactions with special regard to the urogenital tract. Int. J. Med. Microbiol. 295: 9–18
- Servin AL. (2004). Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens.FEMS Microbiol.Rev. 28:405–440.
- FAO/WHO (2002). Guidelines for the evaluation of probiotics in food.Food and Agriculture Organization of the United Nations and World Health Organization Working Group Report.
- Conway PL, Gorbach SL, Goldin BR. (1987) Survival of lactic acid bacteria in the human stomach and adhesion to intestinal cells. J. Dairy Sci. 70(1):1–12.
- Harrigan, W.F. and McCance, M.E., (1990) Laboratory Methods in Food and Dairy Microbiology, 8th edn. Academic Press, London.
- Lee YK, and Puong KY (2002) Competition for adhesion between probiotics and human gastrointestinal pathogens in the presence of carbohydrate. Br. J. Nutr. 88(1): 101–108
- Lahteinen T, Malinen E, KoortJMK.,Hannus UM, Hankimo T, “et al.” (2009) Probiotic properties of Lactobacillus isolates originating from porcine intestine and feces. Anaerobe. 1–8
- Lu Z, Breidt F, Fleming HP, Altermann E and Klaenhammer TR (2003) Isolation and characterization of a Lactobacillus plantarum bacteriophage, AJL-1, from a cucumber fermentation. Int. J. Food Microbiol.84, 225– 235.
- Collins JK, Thornton G, Sullivan GO (1998). Selection of probiotic strains for human applications. Int. Dairy J. 8: 487-490.
- Collins CH, and Lyne PM (1987) Microbiological Methods. Butterwoths. London.
- Mack DR, Ahrne S, Hyde L, Wei S, Hollingsworth MA (2003) Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 52:827–833.
- Maragkoudakis PA, Zoumpopouloua G, Miarisa C, Kalantzopoulosa G, Potb B. “et al.” (2006) Probiotic potential of Lactobacillus strains isolated from dairy products. . Int. Dairy J. 16: 189–199
- Salminen S, Bouley C, Boutron-Ruault, MC, Cummings JH, Franck A, “et al.” (1998). Functional food science and gastrointestinal physiology and function. Br. J. Nutr. 80: 147–171.
- Ouwehand AC, Salminen S, Isolauri E (2002) Probiotics: an overview of beneficial effects. Antonie Van Leeuwenhoek 82: 279–289.
- Chauvière G, Coconnier MH, Kernéis S, Fourniat J, Servin AL. (1992). Adhesion of human Lactobacillus acidophilus strain LB to human enterocyte-like Caco-2 cells. J. Genetic Microbiol. 8: 1689–1696.
- Schiffrin EJ, Rochar F, Link-Amster H (1995) Immunomodulation of human blood cells following the ingestion of lactic acid bacteria. J. Dairy Sci. 78:491–497
- Falagas ME, Rafailidis PI, Makris GC (2008) Bacterial interference for the prevention and treatment of infections. Int. J. Antimicrob. Agents 31: 518–522
- Parkes GC, Sanderson JD, Whelan K (2009) The mechanisms and efficacy of probiotics in the prevention of Clostridium difficile-associated diarrhoea. The Lancet Infect. Dis. 9: 237–244.
- Haza AI, Zabala A, Morales P (2004) Protective effect and cytokine production of a Lactobacillus plantarum strain isolated fromewes’ milk cheese. Int. Dairy J. 14: 29–38
- Kumar H, Rangrez AY, Dayananda KM, Atre1 AN, Patole1MS “et al.” (2011) Lactobacillus plantarum (VR1) isolated from an Ayurvedic medicine (Kutajarista) ameliorates in vitro cellular damage caused by AeromonasVeronii. BMC Microbiolo. 11:152
- Schillinger U, Guigas C, Holzapfel WH (2005) In vitro adherence and other properties of lactobacilli used in probiotic yoghurt-like products. Int. Dairy J. 15: 1289–1297.
- Bernet MF, Brassart D, Neeser JR, Servin AL (1994) Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut. 35: 483-489
- Chan RCY, Bruce AW, Reid G (1984) Adherence of cervical, vaginal and distal urethral normal microbial flora to human uroepithelial cells and the inhibition of adherence of gram-negative uropathogens by competitive exclusion. J. Uro. 131: 596– 601.
- Redondo-López V, Cook RL, Sobel JD (1990) Emerging role of lactobacilli in the control and maintenance of the vaginal bacteria microflora. Rev. Infect. Dis. 12: 856–872.
- Boris S, Suárez JE, Vázquez F, Barbés C (1998) Adherence of human vaginal lactobacilli to epithelial vaginal cells and interaction with uropathogens. Infect. Immun. 66, 1985–1989.
- Tuomola EM, Salminen SJ (1998) Adhesion of some probiotic and dairy Lactobacillus strains to Caco-2 cell cultures. Int. J. Food Microbiol. 41: 45–51.
- Ouwehand AC, Kirjavainen PV, Grönlund MM, Isolauri E, Salminen SJ (1999) Adhesion of probiotic micro-organisms to intestinal mucus. Int. Dairy J. 9: 623–630.
- Rojas M, Ascencio F, Conway PL (2002) Purification and characterization of a surface protein from Lactobacillus fermentum 104R that binds to porcine small intestinal mucus and gastric mucin. Appl. Environ. Microbiol. 68: 2330–2336.
- Buck BL. Altermann E, Svingerud T, Klaenhammer TR (2005) Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71: 8344–8351.
- Pretzer G, Snel J, Molenaar D, Wiersma A, Bron PA “et al.” (2005) Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bactiol.187: 6128–6136.
- Vesterlund S, Paltta J, Karp M, Ouwehand AC (2005) Adhesion of bacteria to resected human colonic tissue: Quantitative analysis of bacterial adhesion and viability. Microbiol. Res. 156, 238–244
- Miyoshi Y, Okada S, Uchimura T, Satoh EA (2006) Mucus adhesion promoting protein, MapA, mediates the adhesion of Lactobacillus reuteri to Caco-2 human intestinal epithelial cells. Biosci.Biotechnol.Biochem. 70: 1622–1628.
- Provencio MD, Llopis M, Antolín M, de Torres I, Guarner “et al.” (2009) Adhesion properties of Lactobacillus casei strains to resected intestinal fragments and components of the extracellular matrix. Arch Microbiol. 191: 153–161.
- Rodríguez M, Zagorec M, Ascencio F, Vázquez-Juárez R, Rojas M (2009) Lactobacillus fermentum BCS87 expresses mucus- and mucin-binding proteins on the cell surface. J. Appl. Microbiol. 2009, 107, 1866–1874.
- Kleerebezem M, Hols P, Bernard E, Rolain T, Zhou M “et al.” (2010) The extracellular biology of the lactobacilli. FEMS Microbiol.Rev. 34: 199–230.
- Coconnier MH, Lie´vin V, Camard MFB, Hudault S, Servin AL (1997) Antibacterial Effect of the Adhering Human Lactobacillus acidophilus Strain LB. Antimicrob. agents and chemotherapy. 1046–1052
- Klaenhammer TR (1993) Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12: 39–85
- Gueimonde ML, Jalonen F, Hiramatsu M, Salminen S (2006) Adhesion and competitive inhibition and displacement of human enteropathogens by selected lactobacilli. Food Res. Int. 39: 467–471.
- Cenic A, Langerholc T, (2010) Functional cell models of the gut and their applications in food microbiology. Int. J. Food Microbiol. 141: 4–14
- McGroarty JA (1994) Cell surface appendages of lactobacilli. FEMS Microbiol.Lett. 124: 405–410.
- Reid, G., McGroarty, J.A., Tomeczek, L., Bruce, A.W. (1996). Identification and plasmid profiles of Lactobacillus species form the vagina of 100 healthy women. FEMS Immun. Med. Microbiol. 15, 23–26.
- Sillanpää J, Martínez B, Antikainen J, Toba T, Kalkkinen N “et al.” (2000) Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J. Bacteriol. 182: 6440-6450.
- Ouwehand AC, Derrien M, de Vos W, Tiihonen K, Rautonen N (2005) Prebiotics and other microbial substrates for gut functionality. Curr.Opin.Biotech. 16(2): 212–217.
- Dunne C, Mahony L, Murphy L, Thornton G, Morrissey D”et al.” (2001) In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am. J. Clin. Nutr. 73: 386-392.
- Tuomola E, Crittenden R, Playne M, Isolauri E, Salminen S (2001) Quality assurance criteria for probiotic bacteria. Am. J. Clin. Nutr. 73: 393–398.
- Charteris, W. P., Kelly, P. M., Morelli, L., & Collins, J. K. (1998). Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J. Appl. Microbiol. 84: 759–768.
- Du Toit M, Franz CM, Dicks LMT, Shillinger U, Haberer P “et al.” (1998) Characterization and selection of probiotic lactobacilli for a preliminary mini pig feeding trial and their effect on serum cholesterol levels, feces pH and feces moisture content. Int. J. Food Microbiol. 40: 93–104.
- Jacobsen, C. N., Roesnfeldt Nielsen, A. E., Moller, P. L., Michaelsen, K. F., Paerregaard, A. et al.(1999). Screening of probiotic activities of forty-seven strains of Lactobacillus sp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 65: 4949–4956.
- Haller D, Colbus H, Ganzle MG, Scherenbacher P, Bode C „et al.“ (2001) Metabolic and functional properties of lactic acid bacteria in the gastro-intestinal ecosystem: A comparative in vitro study between bacteria of intestinal and fermented food origin. Sys. Appl. Microbiol. 24: 218–226.
- Fernandez MF, Boris S, Barbes C (2003) Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J. Appl. Microbiol. 94: 449–455.
- Ouwehand, AC, Tuomola EM, Tolkko S, Salminen S (2001) Assessment of adhesion properties of novel probiotic strains to human intestinal mucus. Int. J. Food Microbiol. 64: 119–126.
- MinelliBE, Benini A, Marzotto M, Sbarbati A, Ruzzenente “et al.” (2004). Assessment of novel probiotic Lactobacillus casei strains for the production of functional dairy foods. Int. Dairy J. 14: 723–736.
- Bron PA, Grangette C, Mercenier A, de Vos WM, Kleerebezem M (2004) Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186(17): 5721–5729.
- De Smet I, Hoorde VL., Woestyne VM., Christiaens H, Verstrate W (1995) Significance of bile salt hydrolytic activities of lactobacilli. J. Appl. Bacteriol. 79: 292–301







