Introduction
Human serum albumin (HSA) is an abundant plasma protein, which attracts great interest in the pharmaceutical industry since it can bind a remarkable variety of drugs impacting their delivery and efficacy and ultimately altering the drug’s pharmacokinetic and pharmacodynamic properties. HSA being the most abundant protein of blood plasma, and has a limited number of high-affinity binding sites.
HSA comprises three homologous domains that assemble to form a heart-shaped molecule. Each domain is a product of two subdomains that possess common structural motifs. The principal regions of ligand binding to human serum albumin are located in hydrophobic cavities in subdomains IIA and IIIA, which exhibit similar chemistry. The structure explains numerous physical phenomena and should provide insight into future pharmacokinetic and genetically engineered therapeutic applications of serum albumin [1].
The three-dimensional structure of human serum albumin has been determined crystallographically to a resolution of 2.8 Å. the structural information of the affinity binding sites is useful when designing new drugs whether the aim is to avoid binding to HSA, or to make use of its depot function [2]. Sudlow et al. did pioneering studies in this field by using a fluorescent probe displacement method in 1975 and 1976 [3,4]. With this method they showed through a screening, that there are two specific drug binding sites on HSA, namely, site I (also called the warfarin binding site) and site II (the benzodiazepine binding site).
Current crystallographic studies have proved that the majority of drugs bind to two main binding sites of high and low affinities [5-8].
Yang et al. proposed that interactive association of drugs binding to HSA usually occurs in site I of HSA, namely, II A subdomain . Site I is a big hydrophobic cavity that is possible to hold several drugs at the same time [9,10]. Site 1 is a preformed binding pocket within the core of subdomain IIA, which comprises six helices of the subdomain and a loop-helix feature (residues 148– 154). The interior of the pocket is hydrophobic, predominantly delimited by residues Trp214, Leu219, Phe223, Leu238, His242, Leu260, Ile264, Ser287, Ile290, and Ala291. However, it also contains two clusters of polar residues, an inner cluster of residues toward the bottom of the pocket (Tyr150, His242, Arg257) and an outer cluster at the pocket entrance composed of Lys195, Lys199, Arg218, and Arg222 (Figure 1). The large binding cavity is comprised of a central zone from which three distinct compartments extend [9].
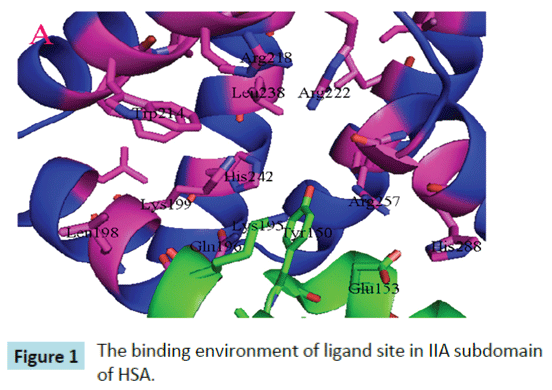
Figure 1: The binding environment of ligand site in IIA subdomain of HSA.
On the other hand, drugs have two forms in circulation, namely, bound or unbound to plasma proteins. The unbound drugs can passively diffuse through the barriers constituted by endothelial cells into the organs where they are metabolized, biliary excretion or glomerular filtration in kidney [11-13]. The unbound drugs can also be distributed intracellularly via specific transport systems. Only free drug molecules interact with therapeutic targets to produce therapeutic effects [11,14]. In most instances, the unbound drug concentration of drug in the tissue depends on the unbound drug concentration in the plasma [11]. Thus, HSA–drug interactions represent an important factor to understand the pharmacokinetics and pharmacological effects of drugs [15-19].
Many drugs and other bioactive small molecules bind reversibly to albumin and other serum components, which then function as carriers. Serum albumin often increases the apparent solubility of hydrophobic drugs in plasma and modulates their delivery to cell in vivo and in vitro [20].
Diclofenac (DIC)
2-(2,6-dichloroanilino) phenylacetic acid (Figure 2a) is a nonsteroidal- anti-inflammatory-drug that is used as an active compound in analgesic, antipyretic, anti-rheumatic medicaments. It is used in the form of sodium or potassium salt [20].
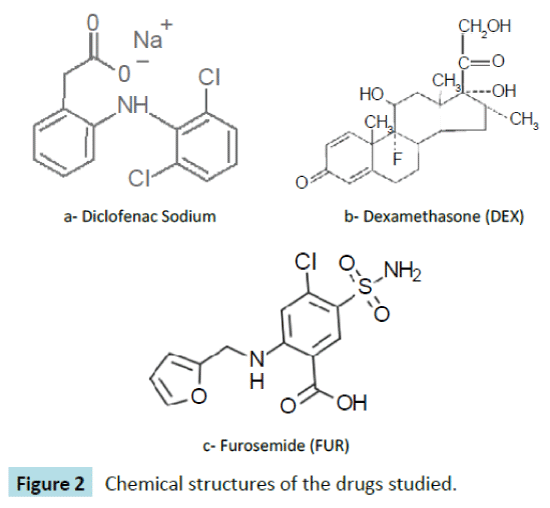
Figure 2: Chemical structures of the drugs studied.
Dexamethasone (DEX)
11b,16a)-9-Fluoro-11,17,21-trihydroxy-16-methylpregna-1,4- diene-3,20-dione (Figure 2b) is a potent synthetic member of the glucocorticoid class of steroid hormones. It acts as an antiinflammatory and immunosuppressant. Its potency is about 20 – 30 times that of hydrocortisone and 4–5 times of prednisone. It is a synthesized corticosteroid compound that has been widely used in clinical treatment for Addison’s disease, systemic lupus erythematosus, periarteritis nodosa and other autoimmune disease [21].
Furosemide (FUR)
4-Chloro-N-furfuryl-5-sulfamoylanthranilic acid (Figure 2c) is primarily used for the treatment of hypertension and edema. It is the first-line agent in most people with edema caused by congestive heart failure. It is also used for hepatic cirrhosis, renal impairment, nephrotic syndrome, in adjunct therapy for cerebral or pulmonary edema where rapid diuresis is required (IV injection), and in the management of severe hypercalcemia in combination with adequate rehydration [22].
In the present study, we focused on the interaction of three widely used drugs (Diclofenac sodium, dexamethasone and furosemide) with HSA in vitro at different temperatures (20-40°C) at pH 7.0 and at different pHs (6.0-8.0) at the same temperature (25°C), via spectrofluorimetric method. Consequently, we made an analysis and interpretation of the fluorescence data. The accurate and comprehensive data accumulated here may help to elucidate the binding mechanism between these drugs and HSA. It may contribute to an enhanced understanding of the transportation, distribution, metabolism, and drug efficacy of these drugs in blood. Also, it may play valuable insights in binding parameters, such as the binding constant, number of binding sites, binding force, and binding distance between the interacting species.
Materials and Methods
Pure drug samples were obtained as gift from the faculty of medicine at the Islamic University of Gaza. HSA (<0.05 % fatty acid) was purchased from Sigma Chemical Co. (St. Louis, MO). All other substances used were of analytical grade and were used as received without further purification. HSA solutions were prepared in the phosphate buffer solutions (20 mM) of the desired pH (6.0 - 8.0) and stored in the dark at 4°C. The pH was checked with a digital pH meter (RIDAO PHS-2C). All reagents used were of analytical reagent grade and distilled water was used throughout the experiments.
Human serum albumin stock solutions (in phosphate buffer solution at the desired pH) were prepared directly prior to experiment and the final concentration of 15 μM was confirmed by absorbance measurement using the absorption coefficient at maximum wavelength (ε278 = 37000 M–1cm–1) and then diluted to 1.5 μM using the same buffer solution. After addition of each drug sample the fluorescence signals were measured within 40 seconds, separated by a 20 s break for the next sample injection.
Drug- HSA interaction
In order to determine the binding parameters of the drugs with HSA, 2 mL of 1.5 μM HSA solution was taken in a quartz cuvette. The bandwidth for measuring emission was 5 nm. The temperature of sample was kept by recycled water throughout the experiment. Then, a successive 20 μL injections of the drug stock solution of 50 μM were added to protein solution. The final drug concentration was in the range of 0 - 0.42 μM. Fluorescence spectra were recorded in the range of 300 - 500 nm after excitation at 278 nm in each case. Intrinsic fluorescence of HSA was measured at 340 nm. Fluorescence spectra were measured with a Perkin Elmer LS50B spectrofluorometer interfaced to a computer for data collection and analysis, using 10 nm/10 nm slit widths. The fluorescence emission spectra of HSA in the absence and presence of increasing amounts of the drugs under study were recorded. All experiments at four different temperatures (in the range of 20-45°C) were performed at pH 7.0. The sample temperature was maintained by recycling water from a super thermostatic water tank (SYC-15) throughout the experiments.
Data analysis
The association constant describing the ligand – protein bond strength was commonly calculated from curve fitting to experimental points. Some researchers fit the equation described by Stern-Volmer to experimentally obtained values of albumin fluorescence and from this determine the affinity constant. The fluorescence quenching data occurred through the drug binding with HSA at different temperatures and at different pH were analyzed according to the modified Stern–Volmer equation [23,24]
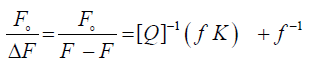 (1)
(1)
Where Fo and F are the relative fluorescence intensities of the protein in the absence and presence of the quencher drug, respectively, ΔF is the difference in fluorescence intensity of Fo and F, f is the fractional maximum fluorescence intensity and K is the effective quenching constant for the accessible fluorophores, which is analogous to the associative binding constant (Kass) for a quencher–acceptor system. The modified Stern-Volmer plots for the effect of temperature change have been shown in Figure 3. A good linear relationship between 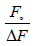 and [Q]−1 was observed, and the slopes were decreased with temperature, and the association constants were inversely correlated with temperature. The results assuming that the quenching process was static.
and [Q]−1 was observed, and the slopes were decreased with temperature, and the association constants were inversely correlated with temperature. The results assuming that the quenching process was static.
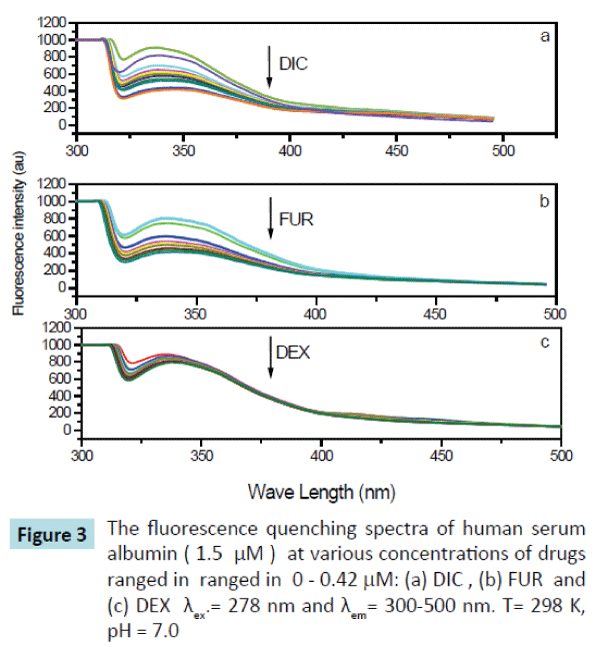
Figure 3: The fluorescence quenching spectra of human serum albumin ( 1.5 μM ) at various concentrations of drugs ranged in ranged in 0 - 0.42 mM: (a) DIC , (b) FUR and (c) DEX λex.= 278 nm and λem= 300-500 nm. T= 298 K, pH = 7.0
Additionally, the fluorescence quenching data were analyzed at different pHs (6.0 -8.0) at the same temperature (25°C), and the corresponding association constants for the three studied drugs were presented in Table 1.
| Drug |
E |
J (cm-3M-1) |
R0 (nm) |
r (nm) |
| Diclofenac sodium |
0.56 |
3.7011x10-14 |
3.1 |
2.98 |
| Furosemide |
0.26 |
1.0772x10-13 |
3.7 |
3.52 |
| Dexamthasone |
0.20 |
2.2468x10-13 |
4.2 |
5.30 |
Table 1: Energy transfer parameters for the interaction of drugs with human serum albumin.
Thermodynamic parameters (ΔG°, ΔH°, and ΔS°) for drug-HSA interaction were determined for the three studied drugs at pH 7.0 at four different temperatures (20, 25, 30 and 40°C).
For both effects using the equations:
ΔG° = - 2.303 RT log Kass. (2)
log Kass = - ΔH°/ 2.303RT + ΔS°/ 2.303 R (3)
ΔG° = ΔH°- TΔS° (4)
The corresponding linear regression equations and results at four different temperatures (20-40°C) are shown in Figure 4 and Table 2. The results showed that the binding constants between the studied drugs and HSA were decreased with temperature, resulting in a reduction of the stability of the drug–HSA complex formed.
| Temperature, K |
Diclofenac Sodium (DIC) |
Stern-Volmer constant (KSV) × 10-4, M-1
Furosemide (FUR) |
Dexamethasone (DEX) |
| 293 |
4.14 |
3.32 |
3.10 |
| 298 |
3.37 |
2.71 |
2.50 |
| 303 |
1.95 |
1.87 |
1.74 |
| 313 |
1.66 |
1.46 |
1.03 |
Table 2: Stern-Volmer binding constant Ksv for the HSA-drug system at pH 7.0 and at different temperatures; λex = 278 nm, λem = 250 – 500 nm.
The spectral overlap curves as well as energy transfer parameter data were imported through Excel spreadsheet into MATLAB R 213a program and then analyzed and visualized as shown in results and discussion. The imported measurements were integrated into an automated analysis workflow in MATLAB program using a written script to calculate the needed parameters.
Results and Discussion
The fluorescence spectra of HSA upon addition of drugs (DIC, FUR and DEX) used in this study were determined and have been shown in Figure 3. It is obvious that HSA fluorescence emission that is peaked at 340 nm was quenched after being excited with a wavelength of 278 nm as the drug concentrations increased. and there was a slight red shift at the maximum wavelength from 340 to 344 nm, which suggested that there was interaction between the drug used and HSA forming drug-HSA complex, making a slight change in the environment around the chromophore of HSA and indicates that a slight conformational change may take place during the binding of the drugs with the HSA. Addition of the drug in question (DIC, FUR, and DEX) leads to quench the fluorescence emission spectra of HSA at 340 nm by variable extents, being the maximum quenching on addition of Diclofenac sodium drug.
The binding constants (Kass) were calculated using the modified Stern–Volmer equation (equation 1) and the results were reported in Table 1. It was found that the interaction between the studied drugs (DIC, FUR and DEX) with HSA occur with variable affinities and the binding constants (Kass) for the studied drugs were decreased with increasing temperature as shown in Figure 4, confirming occurance of static quenching mechanisms in these interactions.
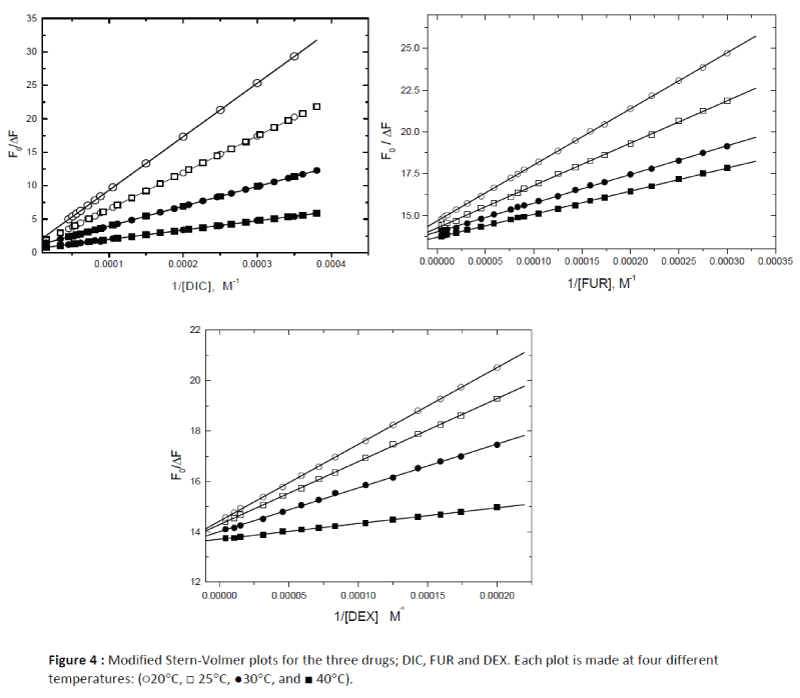
Figure 4: Modified Stern-Volmer plots for the three drugs; DIC, FUR and DEX. Each plot is made at four different temperatures: (?20°C, ? 25°C, ?30°C, and ? 40°C).
Binding constants and effect of temperature
Using the fluorescence quenching data at different temperatures (20 - 40 °C) and at different pHs (6.0 – 8.0) the binding constants of the modified Stern-Volmer model analogous to association constants (Kass) for the binary complexes of each bound drug with HSA were calculated and listed in Table 2.
The association constants obtained in this study indicate that the affinity of these drugs toward HSA is similar to the affinity of other drugs and endogenous substances [25-27].
The decreasing trend of the association constants Kass with increasing temperature was in accordance with the KSV dependence on temperature, resulting in a reduction of the stability of the drug- HSA complex.
The binding constants for the studied drugs with HSA were in the order of: KDIC > KFUR > KDEX. The affinity order for the binding constants among the three drugs studied towards HSA at all temperatures studied were explained by two factors acting together, firstly, the presence of carboxyl group in the drug chemical structure (for DIC and FUR) which is considered as the most reactive species toward albumin. This result was in agreement with previous findings for other drugs and endogenous substances that contain –COOH groups [25-27].
Secondly, the results can be explained on basis of n-octnol/water partition coefficient for these drugs which also follows the order : DIC > FUR > DEX. Hence, diclofenac sodium drug being the most hydrophobic molecule can penetrate the hydrophobic part of subdomain IIA in HSA more easily than the other two drugs.
Stern-Volmer constants which are analogous to binding constants for drug/HSA were calculated according to modified Stern- Volmer (equation 1).
The main HSA binding regions are located in subdomains IIA and IIIA as it results from crystallograghy study of X-ray diffraction for several ligands.
The two subdomains (IIA and IIIA) in HSA have some structural differences that cause the drugs bind somewhat differently in the hydrophobic loop of the subdomain AII in HSA. The portion of the subdomain IIA that binds the hydrophobic part of the drug contains Trp 214 which plays the key role as a chromophore in such binding.
On the other hand, the dexamethasone drug (DEX) as a steroidal drug being a hydrophobic molecule can penetrate the hydrophobic subdomain IIA (in HSA) into which the Trp 214 is located and showed the quenching effect to a lesser extent since it does not contain a carboxylic group as the previous drugs DIC and FUR.
Thermodynamic parameters and nature of the binding forces
The human serum albumin fluorescence quenched due to increasing concentrations of the three drugs studied can indicate that the drugs interact with albumin in the surrounding of the albumin fluorescent residue tryptophan 214 localized in subdomain IIA. The binding of the drug molecule to albumin can affect its fluorescence by acting as a tryptophan quencher via collision between the two targets or via energy transfer.
Generally, there are four types of noncovalent interactions that could play a major role in ligand binding to proteins. These are hydrogen bonds, van der Waals forces, electrostatic, and hydrophobic bond interactions [28]. The binding of the drugs in this study was accompanied by negative enthalpic and negative entropic contributions indicating that van der Waals interactions and hydrogen bonds played the major roles in the binding reaction [29].
Evaluation of thermodynamic parameters for each drug (DIC, FUR and DEX) with HSA, was carried out using equations 2-4 (as written in data and analysis).
Inspection for the values in Table 2 reflects a decrease in the values of binding constant with temperature in the studied range. This thermodynamic stabilization of the drug-HSA complex with the increase in temperature can better be explained by the likely overall conformational whole structure of the conjugate formed rather than the additional local interaction.
The values of ΔH° and ΔS° were calculated from the slope and intercept of the plot of 1n Kass versus 1/T, (respectively). The negative values of both ΔH° and ΔS° indicate that drug interactions with HSA were enthalpic driven and entropic unfavorable. The high negative enthalpy change value for the drug–HSA complex was just as the consequence of the interaction between the two negatively charged species at pH studied.
The association constants obtained at different temperatures (20 – 40 ΔH°C) were treated thermodynamically to calculate the thermodynamic parameters (ΔG°ΔH°, and ΔS°for the studied drug- HSA binding using equations 2-4.
The negative values of ΔG° indicate that drug-HSA complexation is a spontaneous process. Thus the overall stability of drug-DNA complex is dictated by Gibbs free energy change (ΔG°) that involves both enthalpic and entropic contributions.
As seen from Table 3, both enthalpy and entropy changes are negative suggested that Hydrogen bonding and van der Waals forces are the predominant forces that contribute to Gibbs free energy of the drug-HSA interaction.
| Thermodynamic parameter |
Diclofenac sodium |
Furosemide |
Dexamethasone |
| DG° (kJmol-1) |
-25.83 |
-25.29 |
-25.09 |
| DH° (kJmol-1) |
-36.56 |
-32.10 |
-29.06 |
| DS° (Jmol-1) |
-36.66 |
-23.11 |
-15.80 |
Table 3 Thermodynamic parameters for the interaction of drugs (DIC, FUR and DEX)with human serum albumin
Effect of pH on drug binding to HSA
The binding study of different concentrations of the studied drugs and the protein were performed at constant temperature (25°C) in phosphate buffer solutions of different pH values (6.0 – 8.0). A series of successive additions was made for selected drug to HSA as a function of pH and the data are presented in Table 4. The association constant at each pH was calculated as described before using the modified Stern–Volmer equation (equation 1).
| pH |
KDIC × 10-4 |
KFUR × 10-4 |
KDEX × 10-4 |
| 6.0 |
4.25 |
3.76 |
2.81 |
| 6.5 |
3.55 |
3.08 |
2.45 |
| 7.0 |
3.37 |
2.71 |
2.50 |
| 7.5 |
2.14 |
1.45 |
2.37 |
| 8.0 |
1.70 |
0.89 |
2.19 |
Table 4: Binding constants for Drug / HSA interaction at different pHs (6 -8)
in phosphate buffer solution and at 25°C.
In the pH range at which the drug-HSA interaction was carried out, It is believed that both the donor and acceptor species in HSA and drug, respectively were in the anionic forms. The results suggest that the drug molecules become obstructed to penetrate the binding site in the subdomain IIA due to repulsion occurred.
The energy transfer between the drug and HSA
The importance of the energy transfer in biochemistry is that the efficiency of transfer can be used to evaluate the distance between the ligand and the tryptophan residues in the protein [30]. The fluorescence resonance energy transfer model (FRET) was used to determine the distance between the donor, Trp-214 residue in HSA and the acceptor bound drug. The efficiency of FRET model depends mainly on some factors which include: (i) the extent of spectral overlap between the donor emission and the acceptor absorption spectra, (ii) the orientation of the transition dipole of donor and acceptor species and (iii) the distance between the donor and acceptor species.
The distance between the donor and the acceptor can be calculated using FRET model. In which the efficiency of energy transfer, E, can be calculated by Equation 5 :
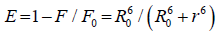 (5)
(5)
Where F and F0 are the relative fluorescence intensities of the protein in presence and the absence of the drug, respectively, R0 is the distance at which the transfer efficiency equals 50%, and r is the distance between donor and acceptor.
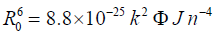 (6)
(6)
R0 is given by the following equation:
Where n is the refractive index of the medium (n=1.36), k2 is the orientation factor (k2=2/3), φ is the quantum yield of the donor (φ = 0.14) [31] and J is the overlap integral between the donor emission and acceptor absorption spectra and is given by the following summation:
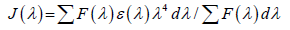 (7)
(7)
The fluorescence quenching studies proved that HSA can form drug-protein complex, and suggest that the energy transfer between the studied drug and HSA occurred with various extents. The energy transfer effect is related not only to the distance between the acceptor and the donor, but also to the critical energy transfer distance.
The values of J, R0, and r are given in Table 1 and the emission spectra for HSA and the absorption spectra of the studied drugs (DIC, FUR and DEX) under the experimental conditions are shown in Figure 5. The comparable values of r and R0 were in accordance with conditions of Förster's non-radiative energy transfer theory and indicated that the energy transfer from donor residue to acceptor drug occurs with high possibility.
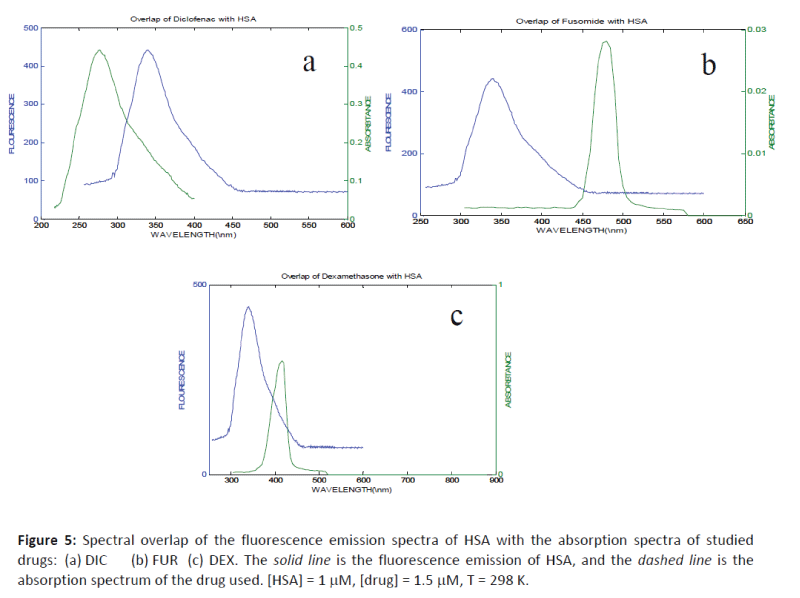
Figure 5: Spectral overlap of the fluorescence emission spectra of HSA with the absorption spectra of studied drugs: (a) DIC (b) FUR (c) DEX. The solid line is the fluorescence emission of HSA, and the dashed line is the absorption spectrum of the drug used. [HSA] = 1 mM, [drug] = 1.5 mM, T = 298 K.
The average distance of r between the donor fluorophore and the acceptor is less than 7 nm, and the fulfillment of the required condition 0.5 R0 < r <1.5 R0 suggests that the energy transfer from HSA to the drug used can occur with high probability. An average r distance on the 2-8 nm scale indicates that the energy transfer occurs with high probability [32-35] and in accordance with the prediction by Förster’s non-radiative energy.
Conclusion
In this paper, we investigated the interaction between three drugs (DIC, FUR and DEX) with HSA by fluorescence spectroscopy. The experimental results indicate that the probable quenching mechanism of fluorescence of HSA by these drugs is static quenching mechanism. The drugs can interact with HSA.
Diclofenac sodium drug bound more effectively than the other two drugs (FUR and DEX). Our results clarified that these drugs can interact with HSA spontaneously through van der Waals interactions and hydrogen bonds is in the hydrophobic pocket of the specific binding site.
The association constants of these drugs with HSA were in the order of 103 - 105 M-1, being decreased with temperature (20 – 40°C). Hydrogen bonding and hydrophobic interactions are involved through the binding process between the studied drugs and HSA. The distances between the drug molecule studied and HSA were between (3-5.5 nm) and were calculated using Förster’s energy transfer theory.
9467
References
- He Min X, Carter CD (1992) Atomic structure and chemistry of human serum albumin. Nature 358: 209-215
- Hein KL,Kragh-Hansen U,Morth JP,Jeppesen MD,Otzen D,et al. (2010) Crystallographic analysis reveals a unique lidocaine binding site on human serum albumin. J Struct Bi 171: 353-360.
- Sudlow G, Birkett DJ, Wade DN (1975) The characterization of two specific drug binding sites on human serum albumin. MolPharmacol 11: 824-832.
- Sudlow G, Birkett DJ, Wade DN (1976) Further characterization of specific drug binding sites on human serum albumin. MolPharmacol 12: 1052-1061.
- Ghuman J, Zunszain PA, Petitpas I, Bhattacharya AA, Otagiri M, et al. (2005) Structural basis of the drug-binding specificity of human serum albumin. J MolBiol 353: 38-52.
- Yang F, Yue J, Ma L, Ma Z, Li M, et al. (2012) Interactive associations of drug-drug and drug-drug-drug with IIA subdomain of human serum albumin. Mol Pharm 9: 3259-3265.
- Curry S (2009) Lessons from the crystallographic analysis of small molecule binding to human serum albumin. Drug MetabPharmacokinet 24: 342-357.
- Curry S (2011) X-ray Crystallography of Albumin. In Human Serum Albumin —New Insights on Its Structural Dynamics, Functional Impacts and Pharmaceutical Applications; University Publishing Center: Kumamoto, Japan 1-29.
- Yang F, Zhang Y, Liang H (2014) Interactive association of drugs binding to human serum albumin. Int J MolSci 15: 3580-3595.
- Simard JR,Zunszain PA, Hamilton JA,Curry S (2006) Location of high and low affinity fatty acid binding sites on human serum albumin revealed by NMR drug-competition analysis. J MolBiol 361: 336-351.
- Yamasaki K, Chuang VT, Maruyama T, Otagiri M (2013) Albumin-drug interaction and its clinical implication. BiochimBiophysActa 1830: 5435-5443.
- Rowland M, Tozer TN, (2010) Clinical Pharmacokinetics and Pharmacodynamics: Concepts and Applications, 4th edition, Lippincott Williams & Wilkins: Philadelphia, PA, USA.
- Koch-Weser J, Sellers EM (1976) Binding of drugs to serum albumin (first of two parts). N Engl J Med 294: 311-316.
- Smith DA, Di L, Kerns EH (2010) The effect of plasma protein binding on in vivo efficacy: misconceptions in drug discovery. Nat Rev Drug Discov 9: 929-939.
- Vallner JJ (1977) Binding of drugs by albumin and plasma protein. J Pharm Sci 66: 447-465.
- Otagiri M (2009) Study on binding of drug to serum protein.YakugakuZasshi 129: 413-425.
- Meyer MC, Guttman DE (1968) The binding of drugs by plasma proteins. J Pharm Sci 57: 895-918.
- Tillement JP, Duché JC, Barré J (2006) [Drug binding to blood proteins: characteristics, roles and pathophysiological changes]. Bull AcadNatl Med 190: 935-946.
- Jusko WJ, Gretch M (1976) Plasma and tissue protein binding of drugs in pharmacokinetics. Drug Metab Rev 5: 43-140.
- Scientific Bulletin of the Technical University of Lodz, (2008). Food Chemistry and Biotechnology72.
- Naik PN,Chimatadar SA,Nandibewoor ST (2010) Interaction between a potent corticosteroid drug–Dexamethasone with bovine serum albumin and human serum albumin: A fluorescence quenching and fourier transformation infrared spectroscopy study. Journal of Photochemistry and Photobiology B: Biology 100: 147-159.
- Sweetman S, Martindale (2009)The complete drug reference. Electronic version. Pharmaceutical Press, Thomson/MICROMEDEX, London, UK/Greenwood Village, Colorado.
- ShuyaCui ,Xiaoli Hu,JiaqinLium (2011) Study of the Binding of Herbacetin to Bovine Serum Albumin by Fluorescence Spectroscopy. J Solution Chem 40: 764-774.
- Lehrer SS (1971) Solute perturbation of protein fluorescence. The quenching of the tryptophyl fluorescence of model compounds and of lysozyme by iodide ion. Biochemistry 10: 3254-3263.
- Levine RL (1977) Fluorescence-quenching studies of the binding of bilirubin to albumin. ClinChem 23: 2292-2301.
- Paál K, Müller J, Hegedûs L (2001) High affinity binding of paclitaxel to human serum albumin. Eur J Biochem 268: 2187-2191.
- Epps DE, Raub TJ, Kézdy FJ (1995) A general, wide-rage spectrofluorometric method for measuring the site-specific affinities of drugs toward human serum albumin. Anal Biochem 227: 342-350.
- Wen MG, Tian JN, Huang YL,Bian HD, Chen ZF,et al. (2009). Interaction between xanthoxylin and bovine serum albumin. Chin. J. Chem. 27: 227-234.
- ZhongW, Wang Y, Yu JS, Liang Y, Ni K, et al. (2004) The interaction of human serum albumin with a novel antidiabetic agent--SU-118. J Pharm Sci 93: 1039-1046.
- Dana S, Iulia M, Carmen M, Mihaela S,Mihaela M,et al, (2009) Spectroscopic Investigations of the Binding Interaction of a New Indanedione Derivative with Human and Bovine Serum Albumins
- Bi S, Song D, Tian Y, Zhou X, Liu Z, et al. (2005) Molecular spectroscopic study on the interaction of tetracyclines with serum albumins. SpectrochimActa A MolBiomolSpectrosc 61: 629-636.
- Yang MM, Yang P, Zhang LW (1994) Study on interaction of caffeic acid series medicine and albumin by fluorescence method. Chin Sci Bull 39: 31.
- Shuang Li, Di Y,Hedong B,Zhenfeng C, Jing Y,et al. (2011). Interaction BetweenPlumbagin and Human Serum Albumin by Fluorescence Spectroscopy. J Solution Chem 40: 709-718.
- Hu YJ, Liu Y, Zhang LX (2005) Study of Interaction BetweenColchicines and Bovine Serum Albumin by Fluorescence Quenching Method. J MolStruct 750: 174-178.
- He W, Li Y, Xue C, Hu Z, Chen X, et al. (2005) Effect of Chinese medicine alpinetin on the structure of human serum albumin. Bioorg Med Chem 13: 1837-1845.







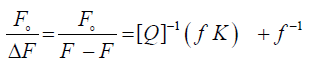 (1)
(1)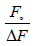 and [Q]−1 was observed, and the slopes were decreased with temperature, and the association constants were inversely correlated with temperature. The results assuming that the quenching process was static.
and [Q]−1 was observed, and the slopes were decreased with temperature, and the association constants were inversely correlated with temperature. The results assuming that the quenching process was static.

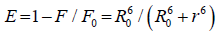 (5)
(5)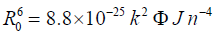 (6)
(6)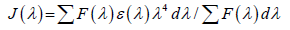 (7)
(7)