Keywords
Pathogens; E. coli
Introduction
Given the specific resistance of bacterial strains to antibiotics, the determination of sensibility to antimicrobial drugs is very important for the monitoring of pathogens and may be used as a necessary epidemiological marker for mentioned bacteria [1-4]. Medications, treatment of the infectious diseases gains more and more problematic characteristics. The reason for this is the fact that throughout the time there have been produced strains of bacteria, which are stable, selective and resistant towards various medication, in the nature. The latter fact has significantly reduced effects of many antibiotics, Sulfanilamide and of other medicines used for treatment, in some cases they even got useless. Resistance towards antibiotics, obtained by microorganisms, remains to be an important problem until today, because of the wide distribution of the resistant forms of bacteria and reduced effectiveness of existing and widely utilized medicament in the sphere of medicine. Poly-resistant strain that cause group infection diseases [5-7]. Treatment of the intestinal infections represents a unification of the complex measures and includes therapy-first of all detoxication. Treatment is focused on the inactivation of the cause of the disease, besides, throughout the treatment the microbial balance, which later on determines normal viability of the organism, should not be disturbed. While selecting the medication, firstly, the sensibility of the cause towards chosen form of treatment should be taken into consideration. According to the standards of the National Committee for Clinical Laboratory Standards (NCCLS), antimicrobial medication may be utilized for the treatments, whenever 80% of the strains demonstrate sensibility towards them [8-10].
The alternative for the antibiotics are commercial phage medicines, among which is Coli Bacteriophage, which contains specific bacteriophages, which has specific bacteriophages against pathogenic E. coli. Using phage-based medications are harmless for human body, since phages have only lysis activity toward pathogenic bacteria [11,12].
The represented work here aims to identify pathogenic E. coli causing intestinal infections in the clinical samples as well as in the food (ground-beef). To define the sensibility of the antibiotics towards the mentioned strains also along with commercial phage medicine-Coli Phage, the structure of which contains bacteriophages against pathogenic E. coli. In addition, to determine sensibility of pathogenic strains towards this medicine.
Materials and Methods
Isolation of E. coli strains
For isolation of E. coli strains, a loopful from the diluted specimens was inoculated into MacConkey's agar and incubated at 37°C for 18-24 hours. Bacterial colonies were identified based on gross morphology, a number of colonies and the hemolytic pattern. Appropriate biochemical and serological tests were performed on the colonies isolated to identify the pathogens.
Serogrouping of E. coli isolates
E. coli serogroups were identified serologically by slide agglutination test using standard polyvalent and monovalent E. coli antisera (O114, O86, O26, O55, O36, O111, O119, O125, O126, O142, O157) according to Edwards and Ewing (1972) [13].
Antimicrobial susceptibility testing
To analyze the anti-biogram profile of the isolates, all isolated E. coli strains were tested for susceptibility to 15 different antimicrobial agents that are most widely used in clinics: penicilin (P), ampicillin (A), carbenicilin (Cb), ampiox (Ap) chloramphenicol (C), streptomycin (S), tetracycline (T), gentamicin (G), kanamycin (K), eritromicin (E), meticilin (M), fortum (F),Cephamezin (Cf), ketotefen (Kt), claforan (Cl) Azythromicin (Az), Ciprofloxacin (Cip), Imipenem (Im), Cefazoline (Cef), Polymoxin (Pm). The susceptibility of these isolates to antimicrobial agents was performed by diskdiffusion method in accordance with the standards of the National Committee for Clinical Laboratory Standards [14].
Identification system of the bacteria of the intestinal group
Represents a standardized system, which contains 21 mini biochemical tests and is applied to identify the bacteria of the gastrointestinal group and other gram-negative bacteria.
Principle
Api 20 E consists of 20 micro test-tubes, which contain dehydrated substrates. Bacterial suspense inputted into stripe gets into reaction with a given substrate. While incubation the color changes, which might be defined directly or through adding the reagents.
The process of reading the reactions is carried out through utilizing respective interpretative table.
While cumulating positive results digital profile is produced and through respective analytical catalogue the name and the variety of cause is defined.
Bacteriophage isolation
The bacteriophages were isolated from wastewater by filtration, followed by the addition of broth concentrate and 18 h cultures of different strains (test strains) to the filtrate. After 24 h incubation in a thermostat, the mixture was filtered through filters with a pore diameter of 0.45 μm (Millipore, United States), and the filtrates were spot-tested for the presence of phages by application of the filtrate (0.1 mL) on the lawn of the test strain on a solid nutrient medium. The result was considered positive if there was a lysis zone on the lawn in 18-24 h of cultivation at 37°? [15]. The highly specific bacteriophage strains were screened and selected by the plate method according to Gracia [16].
Bacteriophage cloning by repetitive single plaque isolation
The phages isolated from the initial material are not uniform. Mixtures of various phage types are frequent, as judged by plaque morphology. Plaques are of various sizes determined by virion size, speed of phage reproduction, etc. Plaques have other characteristic features, such as the sharpness of their edges, the presence of a halo and the completeness of the clearing. A single plaque may contain 107-109 virions. A number of theoretical and practical considerations require the use of “pure” phage lines (clones) during therapy. For this purpose an isolated phage plaque is touched with the fine end of a Pasteur pipette and small specimen is collected, subjected to serial dilution, mixed with plating bacteria and molten soft agar and spread on a plate. Isolation of a reliably pure bacteriophage clone requires about 3-5 such steps.
Electron microscopy: The morphology group membership of bacteriophages was investigated by the method of electron microscopy. Phages with a high titer of >1010 CFU/mL were negatively stained with 1% uranyl acetate. Pictures were obtained by transmission electron microscopy (JEM 100 SX, JEOL, Japan) with negative contrasting of the preparations by uranyl acetate.
Results and Discussion
To reach out the defined objectives and aims, in frames of the project we have communicated with clinical laboratory, from there we received strains of the E. coli derived from the patients. From the mentioned laboratory we got the various E. coli derived from the patients for the further research.
From the Bacteriological Laboratory of the clinical we were given-56 pathogenic strain of E. coli. The mentioned pathogens were derived from the various organs of the patients, as follows: Ear, dis-bacteriosis, pus, wound, skin.
Herewith, from the second Bacteriological Laboratory we were given strains of the pathogenic E. coli. The mentioned pathogens were derived from the various organs of the patients, as follows: Ear, dis-bacteriosis, pus, wound, and skin. From the second bacterial laboratory, we were handed over with 33 strains of the pathogenic E. coli.
In April-May, 2019, the number of various strains given, in accordance with the biosafety procedures, from the clinical microbiological laboratories had been totaled 89.
The above mentioned strains were researched according to the morphological, cultural, biochemical, serological and pathogenic characteristics (Figure 1).
Figure 1: Api systems in E. coli diagnostics.
Based on the specifics of the work we have as well started conducting analysis on microbiological pollution of foods, namely of ground- beef and pork (Figure 2).
Figure 2: Hemolytic activity of E. coli.
In total 20 samples were studied, out of which in 16 cases E. coli were identified.
Investigation of morphological, cultural properties and their antigenic structure of the isolated bacteria indicated that the isolated strains of E. coli belong to the serotypes, such as O114 (30%), O26 (20%), O36 (25%), O125 (15%), O126 (10%).
Pathological strains of E. coli derived from the patients were assessed on the antibiotic resistancy. Some of the strains were resistant towards: Azythromicin, Ciprofloxacin, Imipenem, Cefazolin, Polymoxin and chloramphenicol (Figures 3 and 4). Majority of the strains were resistant towards two and more antibiotics.
Figure 3: Antibioticograms of the various bacterial strains.
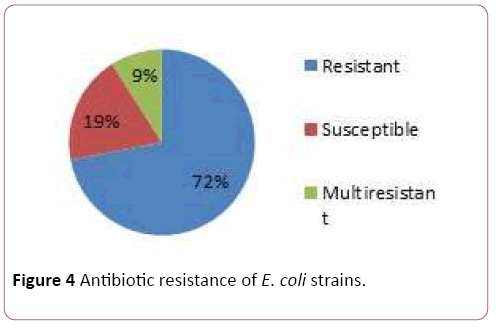
Figure 4: Antibiotic resistance of E. coli strains.
Ground meat bacteriological tests were performed to detect contamination of the above mentioned products.
Overall 20 samples were assessed, out of which 16 samples were contaminated by E. coli. This strains were characterized by resistance towards two or more antibiotics, in some casestowards seven antibiotics.
E. coli strains derived from ground meat were resistant against: Amicacin, Azythromicin, Polymixin, Imipenem. Interesting fact to mention is that strains of E. coli derived from the ground meat are resistant towards the same antibiotics (Amicacin, Azythromicin, Polymixin, Imipenem) as the strains of E. coli derived from the patients in the lab (Figure 5). Antibiotic susceptibility test revealed high resistance (Figure 5).
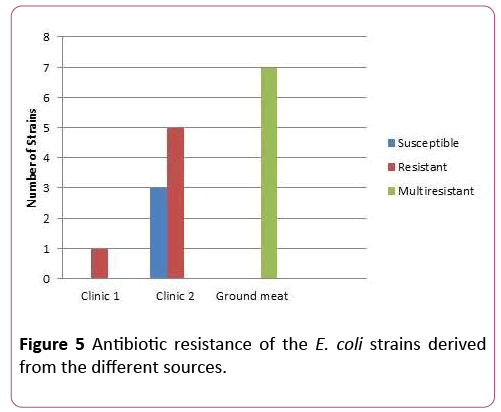
Figure 5: Antibiotic resistance of the E. coli strains derived from the different sources.
22 strains out of 56 of E. coli obtained from clinic I showed resistance toward antibiotics. From 33 E. coli strains obtained from clinic II only 9 isolates revealed resistance. High resistance was shown from strains isolated from ground meat-32 resistant and 7 multi-resistant strains (resistant to four or more antibiotics).
To isolate bacteriophages effective against new isolated pathogenic E. coli strains 40 samples of urban sewage water, river water and other environmental sources were investigated. From them 16 samples contained phages lysed strains of pathogenic E. coli. For formulating polyvalent phage preparation with widest spectrum of action and high lytic activity all 16 phages were characterized according to host range. These phages were tested against all 89 new isolated pathogenic E. coli clinical strains and 16 E. coli strains isolated from food. 6 phages conditionally named vB-Eco1, vB-Eco2, vB-Eco3, vB-Eco4, vB-Eco5 and vB-Eco6, with wide, complementary, non-overlapping host ranges were selected for further characterization (Figures 6-8).
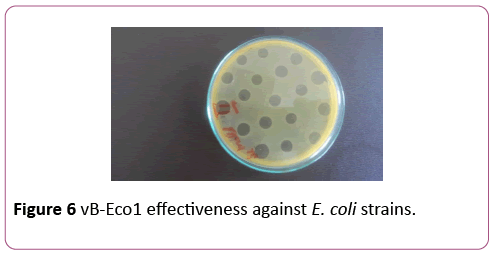
Figure 6: vB-Eco1 effectiveness against E. coli strains.
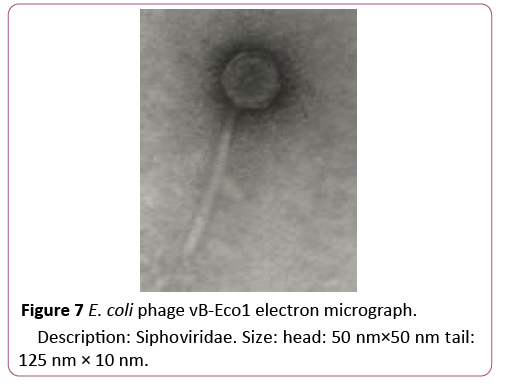
Figure 7: E. coli phage vB-Eco1 electron micrograph.
Description: Siphoviridae. Size: head: 50 nm×50 nm tail: 125 nm × 10 nm.
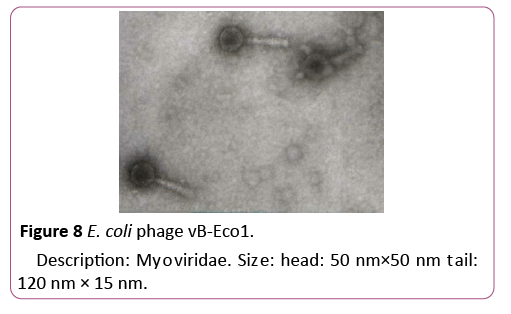
Figure 8: E. coli phage vB-Eco1.
Description: Myoviridae. Size: head: 50 nm×50 nm tail: 120 nm × 15 nm.
A phage preparation on the base of 6 newly isolated and characterized bacteriophages with a high range of activity against different strains of pathogenic E. coli was created. This preparation was effective against 89.9% of investigated strains. The study revealed that 80 out of 89 strains of E. coli were sensitive towards Coli phage cocktail. Also 14 out of 16 strains of E. coli derived from the ground meat were 87.5% sensitive towards Coli phage cocktail (Figure 9).
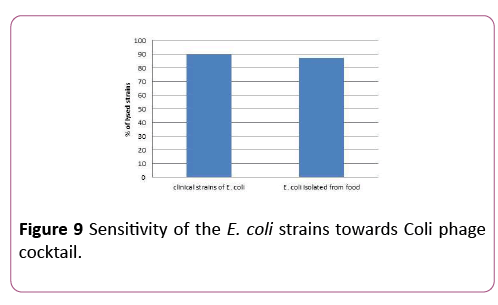
Figure 9: Sensitivity of the E. coli strains towards Coli phage cocktail.
Considering high lytic activity and broad spectrum of this polyvalent phage preparation, it will be significant alternative for prevention and treatment of colibacillosis caused by multidrug resistant E. coli pathogens.
Conclusion
The bacteriological laboratory of the first clinic tested 56 pathogenic strains that were resistant towards: Azythromicin, Ciprofloxacin, Imipenem, Cefazolin, Polymixin and Chloramphenicol.
• The bacteriological laboratory of the second clinic tested 33 pathogenic strains that were resistant towards: Amicacin, Azythromicin, Ciprofloxacin, Imipenem, Polymixin. • 20 samples of ground meat were studied in April-May, 2019. In 16 samples was detected. The strains were resistant towards two or more, in several cases as much as seven antibiotics. Strains derived from ground meat were resistant towards: Amicacin, Azythromicin, Imipenem, Polymixin. • We have produced the phages from the 89 strains derived from the patients and 16 strains derived from the ground meat and created the cocktail with the 6 most effective phages. 89.9% of patient derived strains and 87.5% of meat derived strains were sensitive towards this cocktail. The study was performed in Petre Shotadze Tbilisi Medical Academy and was funded by the academy grant project. Part of the phage phase was performed in G. Eliava Institute of bacteriophages, microbiology and virology.
Acknowledgements
None
Declarations
Funding: None
Conflict of interest: None declared
26405
References
- Kaper JB, Nataro JP, Mobley HLT (2004) Pathogenic Escherichia coli. Nat Rev Microbiol 2: 123-140.
- Tozzoli R, Scheutz F (2014) Diarrhoeagenic Escherichia coli infection in humans. In: Morabito S (ed.) Pathogenic Escherichia coli. Molecular and Cellular Microbiology. 1st ed. Caister Academic Press; Norfolk 1-18.
- Cavaco LM (2009) A novel gene conferring transferable quinolone resistance in Salmonella entericaserovars Kentucky and Bovismorbificans strain of human origin. Antimicrob Agents Chemother 53: 639-645.
- Bonnedahl J, Drobni P, Johansson A (2010) Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum β-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J Antimicrob Chemother 65: 1939-1944.
- Sengupta S, Chattopadhyay MK, Grossart HP (2013) The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol 4: 47.
- Bonnedahl J, Drobni P, Johansson A (2010) Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum β-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J Antimicrob Chemother 65: 1939-1944.
- Spellberg B, Gilbert DN (2014) The future of antibiotics and resistance: a tribute to a career of leadership by John Bartlett. Clin Infect Dis 59: S71-S75.
- Lushniak BD (2014) Antibiotic resistance: a public health crisis. Public Health Rep 129: 314-316.
- Bhoomika SS, Patyal A, Gade NE (2016) Occurrence and characteristics of extended-spectrum β-lactamases producing Escherichia coli in foods of animal origin and human clinical samples in Chhattisgarh, India. Vet World 9: 996.
- Yang C, Lin M, Liao P (2009) Comparison of antimicrobial resistance patterns between clinical and sewage isolates in a regional hospital in Taiwan. Lett Appl Microbiol 48: 560-565.
- Gabisonia T, Chakhunashvili N, Katamadze T, Loladze M, Tamarashvili N, et al. (2018) CHARACTERISTICS OF NEW BACTERIOPHAGES AGAINST THE STRAINS OF ESCHERICHIA COLI. Bulletin of the Georgian National Academy of Sciences 12: 3.
- Hagens S, Loessner MJ (2007) Application of bacteriophages for detection and control of foodborne pathogens. Appl Microbiol Biotechnol 76: 513-519.
- El-Seedy FR, Abed AH, Yanni HA, Abd El-Rahaman SAA (2016) Prevalence of Salmonella and E. coli in neonatal diarrheic calves. J Basic Appl Sciences 5: 45-51.
- Bauer AW, Kirby MM, Sherris JC, Truck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45: 493-496.
- Kutter E, Sulakvelidze A, Raton B (2004) Bacteriophages: Biology and Applications, FL: CRC Press, 2004.
- Adams M, Bakteriofagi (Bacteriophages), Moscow: Inostrannaya literatura, 1961.














