Keywords
Sepsis; Resistance; Antibiotics; Bacteria pattern
Abbreviations:
IZ: Inhibition Zone; S: Sensitive; R: Resistant; AMP: Ampicillin; CN: Gentamicin; LEV: Levofloxacine; VA: Vancomycin; MTZ: Metronidazole; MEM: Meropenem; RD: Rifampicin; CRO: Ceftriaxone; CIP: Ciprofloxacine; E: Erythromycin; MXF: Moxifloxacine; TGC: Tigecycline; LZD: Linezolid; CEZ: Ceftazidime; AK: Amikacin; Sulb-cef: Sulbactam- Cefoperazone; NA: Nutrient Agar; CLSI: Clinical and Laboratory Standards Institute; BHI: Brain Heart Infusion; CFU: Colonyforming Units; TNF: Tumor Necrosis Factor; IL-6: Interleukin 6; CRP: C-reactive Protein; PAMPs: Pathogen-associated Molecular Patterns; DNA: Deoxyribonucleic Acid; CNS: Central Nervous System; MIC: Minimum Inhibitory Concentration.
Background
Infectious can occur because of infection agents such as bacteria, fungus, virus, and protozoa [1]. Bacteremia is state which presence of living bacteria on blood stream [2]. Bacteremia is states that determine to occur sepsis. Further, sepsis can develop to be sepsis shock and also renal and liver dysfunction with hypotension [3]. Bacteria are one of sepsis’s causes. It comes from previous infections such as pulmonary infection, urinary tract infection, central nerve system infection, or Community acquired methicillin-resistant Staphylococcus aureus [3,4]. Sepsis patients will have possibility for developing complications such as organ complications. One of factor in-creasing mortality on sepsis patient was organ dysfunction. Mortality of sepsis patients were estimated about 15% and increased to be 70% if patients suffer three or more organ dysfunction including pulmonary, renal, or heart [5]. Development of bacteria resistance was increasing quickly by discovering resistant bacteria against antibiotics in 1979 – 2011. Some bacteria had been being resistant against antibiotics, for instance gentamicin-R Enterococcus, vancomycin-R Enterococcus, levofloxacin-R Pneumococcus, imipenem-R Enterobacteriaceae, vancomicin-R Staphylococcus, ceftriaxone-R Nesseria gonorrhoeae, and ceftaroline-R Staphylococcus [6]. Research about Escherichia coli resistance showed that 21 bacteria isolates (0,6%) were resistant against ampicillin, chloramphenicol, gentamicin, ciprofloxacine, cefotaxime, and trimethoprim/ sulfamethoxazole [7]. Lewis et al. reported that resistance also occurred against ceftriaxone and imipenem on Acinetobacter spp. (28.6% and 10%), Pseudomonas aeruginosa (46.7% and 3.8%), and Enterobacter spp. (16% and 0%) [8]. Research was performed at ICU room, Fatmawati hospital, Indonesia, revealed that Pseudomonas aeruginosa, Staphylococcus epidermidis, and Escherichia coli were resistant against meropenem, gentamicin, and levofloxacine [9].
Method
Total of bacteria isolates were 53 items consist of 7 bacteria isolates from adult sepsis patients and 46 secondary data of sensitivity test against antibiotics. 7 bacteria isolates would be performed in microbiology laboratory, Faculty of Pharmacy, MUS, to know sensitivity against antibiotics and 46 secondary data of sensitivity test have been performed by microbiology laboratory of Dr. Moewardi general hospital. Bacteria were isolated by standard of microbiology laboratory in Dr. Moewardi general hospital and then obtained a single colony of bacterium. The bacteria isolates were grown on NA (Nutrient Agar) and brought to microbiology laboratory of Faculty of Pharmacy, MUS, for sensitivity test against antibiotics. Sensitivity test was carried out using disc diffusion and the results were interpreted based on CLSI (Clinical and Laboratory Standards Institute). The bacteria isolates, from NA, were taken by using sterile inoculation loop and put into BHI medium then shook by shaker for 2 hours. After that, took 200 μL of BHI solution and diluted by NaCl 0.9% until having similar turbidity with Mc Farland (1.5 × 108 CFU/mL). Bacteria suspension was taken 100 μL then put it on Mueller-Hinton agar and distributed it evenly. The next step, 8 antibiotic discs were put on Mueller-Hinton agar surface and incubated on 37°C for 24 hours (Figure 1).
Figure 1 Sensitivity test result by disc diffusion method on Acinetobacter baumannii. 1: Ceftriaxone; 2: Meropenem; 3: Gentamicin; 4: Metronidazole; 5: Rifampicin; 6:Levofloxacine; 7: Ampicillin; 8: Vancomycin
The inhibitory zone of each antibiotic disc was measured on the next day. Analysis of the result of bacteria sensitivity were determined by measuring inhibition zone of each antibiotic discs and compared by CLSI standard (ampicillin, gentamicin, levofloxacine, vancomycin, metronidazole, meropenem, rifampicin, and ceftriaxone), and coupled by secondary data (sensitivity test data from microbiology laboratory of Dr. Moewardi general hospital). Based on the previous result (merge primary and secondary data), so we could classify level of bacteria sensitivity on adult sepsis, included sensitive (S) or resistant (R). This research is ethically approved by Health Research Ethics Committee Dr. Moewardi General Hospital/ School of Medicine Sebelas Maret University of Surakarta. Ethical Clearance number is 476/ VII/HREC/2014.
Result
This research was carried out to determine bacteria and its resistance pattern on adult sepsis patient against some antibiotics at Dr. Moewardi general hospital in 2014. Patient distribution based on age and gender at Dr. Moewardi general hospital can be seen in Table 1.
| |
Total |
Percentage (%) |
| Gender |
Man |
27 |
58.7 |
| Woman |
19 |
41.3 |
| Total |
46 |
100 |
| |
| Age (year) |
20-30 |
3 |
6.52 |
| 31-40 |
8 |
17.4 |
| 41-49 |
6 |
13.04 |
| >49 |
29 |
63.04 |
| Total |
46 |
100 |
Table 1 Distribution of bacteria isolates based on age and gender at Dr. Moewardi general hospital in 2014.
Based on Table 1, sepsis frequently occurs on patients who older than 49 years old. 7 bacteria isolates include Grampositive and Gram-negative bacteria covering Staphylococcus haemolyticus, Staphylococcus epidermidis, Acinetobacter baumannii, Pseudomonas aeruginosa, and Escherechia coli.
The seven isolates were tested their resistance against eight antibiotics (ampicillin, gentamicin, levofloxacine, vancomycin, metronidazole, meropenem, rifampicin, and ceftriaxone). The results were shown on Tables 2 and 3. In the result, the 7 (Tables 2 and 3) of 53 bacteria were resistant to ampicillin and metronidazole, but sensitive against meropenem. Acinetobacter baumannii showed resistant to some antibiotics such as ampicillin, vancomycin, metronidazole, rifampicin, and ceftriaxone. Resistance was also shown Pseudomonas aeruginosa against ampicillin, vancomycin, metronidazole, and rifampicin. For Acinetobacter baumannii and Pseudomonas aeruginosa were sensitive to gentamicin, levofloxacine, and meropenem whereas Pseudomonas aeruginosa was sensitive to ceftriaxone.
Bacteria
(bacteria code) |
Antibiotics |
| MEM1 |
CN1 |
LEV1 |
VA1 |
MTZ2 |
AMP2 |
RD2 |
CRO2 |
| IZ |
C |
IZ |
C |
IZ |
C |
IZ |
C |
IZ |
C |
IZ |
C |
IZ |
C |
IZ |
C |
Staphylococcus
Haemolyticus(618D) |
8.5 |
R |
9 |
R |
14 |
R |
14.5* |
R |
- |
R |
- |
R |
- |
R |
17 |
R |
Staphylococcus
Epidermidis
(68 D) |
31.7 |
S |
19 |
S |
28.8 |
S |
13.8 |
R |
- |
R |
23 |
R |
12 |
R |
17.5 |
R |
1: Antibiotic on antibiotic guideline for sepsis patient at Dr. Moewardi general hospital; 2: antibiotic on prescription for sepsis patient
* = irradical -: no inhibition zone
IZ: Inhibition Zone (mm); C: Category; S: Sensitive; R: Resistant; AMP: Ampicillin; CN: Gentamicin; LEV: Levofloxacine; VA: Vancomycin; MTZ: Metronidazole; MEM: Meropenem; RD: Rifampicin; CRO: Ceftriaxone |
Table 2 The result of sensitivity test of Gram-positive from adult sepsis patient.
Bacteria
(Bacteria code) |
Antibiotics |
| MEM1 |
CN1 |
LEV1 |
VA1 |
MTZ2 |
AMP2 |
RD2 |
CRO2 |
| IZ |
C |
IZ |
C |
IZ |
C |
IZ |
C |
IZ |
C |
IZ |
C |
IZ |
C |
IZ |
C |
Acinetobacter
Baumannii (625 D) |
25.8 |
S |
16 |
S |
23.5 |
S |
- |
R |
- |
R |
17 |
R |
12.3 |
R |
15 |
R |
| Escherichia coli (646 D) |
22.6 |
S |
19.75* |
R |
23.25* |
R |
11* |
R |
- |
R |
- |
R |
14.7 |
R |
- |
R |
| Escherichia coli (770 D) |
30 |
S |
7.7 |
R |
26.7 |
S |
25* |
R |
- |
R |
- |
R |
21.7 |
S |
14.7 |
R |
| Escherichia coli (866 D) |
30 |
S |
19 |
R |
26 |
R |
21 |
S |
- |
R |
- |
R |
16 |
R |
12.8 |
R |
Pseudomonas
Aeruginosa (790 D) |
4 |
S |
18.8 |
S |
27.8 |
S |
- |
R |
- |
R |
- |
R |
13* |
R |
17.5 |
S |
1: Antibiotic on antibiotic guideline for sepsis patient at Dr. Moewardi general hospital; 2: antibiotic on prescription for sepsis patient
*: irradical; -: no inhibition zone
IZ: Inhibition Zone (mm); C: Category; S: Sensitive; R: Resistant; AMP: Ampicillin; CN: Gentamicin; LEV: Levofloxacine; VA: Vancomycin; MTZ: Metronidazole; MEM: Meropenem; RD: Rifampicin; CRO: Ceftriaxone |
Table 3 The result of sensitivity test of Gram-negative on adult sepsis patient.
Resistance pattern of Gram-positive
29 bacteria strains were obtained from adult sepsis that consist of two isolates (primary data) and 27 isolates (secondary data) (Table 4). Resistance patterns were performed to all of these isolates and the results were shown in Figure 2. Resistance was also found against erythromycin on Staphylococcus epidermidis, Staphylococcus haemolyticus, and Staphylococcus hominis (Figure 2). The test result showed that there were some bacteria that resistant against antibiotics. Staphylococcus aureus and Staphylococcus haemolyticus were resistant to vancomycin.
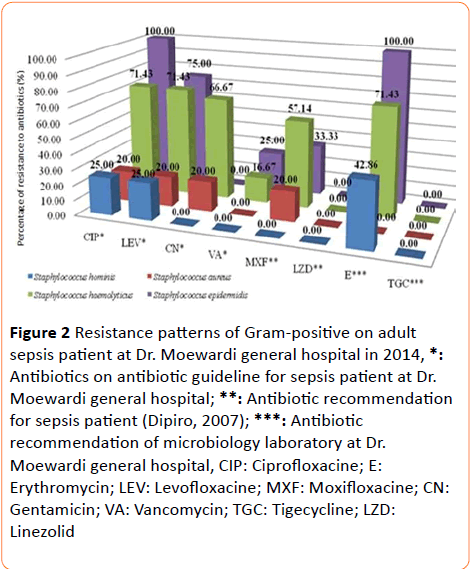
Figure 2: Resistance patterns of Gram-positive on adult sepsis patient at Dr. Moewardi general hospital in 2014, *: Antibiotics on antibiotic guideline for sepsis patient at Dr. Moewardi general hospital; **: Antibiotic recommendation for sepsis patient (Dipiro, 2007); ***: Antibiotic recommendation of microbiology laboratory at Dr. Moewardi general hospital, CIP: Ciprofloxacine; E: Erythromycin; LEV: Levofloxacine; MXF: Moxifloxacine; CN: Gentamicin; VA: Vancomycin; TGC: Tigecycline; LZD: Linezolid
| Bacteria |
Primarydata |
Secondarydata |
Total |
Percentage (%) |
| |
|
| Gram-positive |
|
|
|
|
| Staphylococcus hominis |
|
8 |
8 |
15.09 |
| Staphylococcus haemolyticus |
1 |
7 |
8 |
15.09 |
| Staphylococcus aureus |
|
5 |
5 |
9.43 |
| Staphylococcus epidermidis |
1 |
4 |
5 |
9.43 |
| Staphylococcus capitis |
|
1 |
1 |
1.89 |
| Enterobacter cloceae |
|
1 |
1 |
1.89 |
| Enterococcus faecium |
|
1 |
1 |
1.89 |
| Total |
2 |
27 |
29 |
54.71 |
| Gram-negative |
|
|
|
|
| Escherichia coli |
3 |
4 |
7 |
13.21 |
| Acinetobacter baumannii |
1 |
5 |
6 |
11.32 |
| Klebsiella pneumoniae |
|
6 |
6 |
11.32 |
| Achromobacter xylosoxidant |
|
2 |
2 |
3.77 |
| Proteus mirabilis |
|
1 |
1 |
1.89 |
| Pseudomonas aeruginosa |
1 |
|
1 |
1.89 |
| Salmonella sp. |
|
1 |
1 |
189 |
| Total |
5 |
19 |
24 |
45.29 |
| Total (Gram-positive+ Gram-negative) |
7 |
46 |
53 |
100.00 |
Table 4 Result of bacteria isolates from adult sepsis patients at Dr. Moewardi general hospital in 2014.
On the other hand, Staphylococcus hominis was not resistant against antibiotics that were used as antibiotic recommendation for sepsis, such as gentamicin and vancomycin. All of isolated Gram-positive bacteria (Figure 2) were sensitive against linezolide and tigecycline. Staphylococcus aureus, Staphylococcus hominis, and Staphylococcus epidermidis were sensitive to moxifloxacin, so it can be used as antibiotic for sepsis. Dipiro et al. reported that moxifloxacine and linezolid could be used as empiric antibiotic for sepsis patient [10]. From the statement above, moxifloxacine and linezolid can be used as recommendation empiric antibiotic in sepsis patient. Recommendation empiric antibiotic in sepsis patient.
Resistance pattern of gram-negative
24 gram-negative bacteria were earned from adult sepsis patients consist of 5 bacteria isolates from primary data and 24 isolates from secondary data. Escherichia coli are natural bacteria in colon, but Escherichia coli were found on sepsis patient. Not only Escherichia coli but also Acinetobacter baumannii, Klebsiella pneumoniae, Achromobacter xylosoxidant and Pseudomonas aeruginosa were found on sepsis patient (Figure 3).
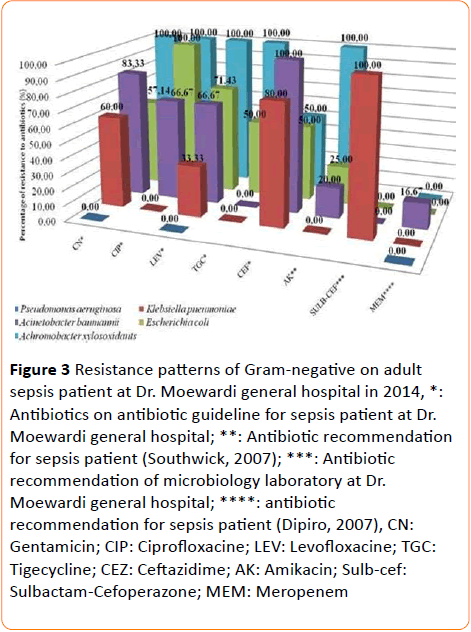
Figure 3 Resistance patterns of Gram-negative on adult sepsis patient at Dr. Moewardi general hospital in 2014, *: Antibiotics on antibiotic guideline for sepsis patient at Dr. Moewardi general hospital; **: Antibiotic recommendation for sepsis patient (Southwick, 2007); ***: Antibiotic recommendation of microbiology laboratory at Dr. Moewardi general hospital; ****: antibiotic recommendation for sepsis patient (Dipiro, 2007), CN:
Gentamicin; CIP: Ciprofloxacine; LEV: Levofloxacine; TGC: Tigecycline; CEZ: Ceftazidime; AK: Amikacin; Sulb-cef: Sulbactam-Cefoperazone; MEM: Meropenem
Based on the research, Klebsiella pneumoniae, Acinetobacter baumannii, and Escherichia coli were resistant against levofloxacine, ciprofloxacine, gentamicin, amikacin. Otherwise, all of bacteria were sensitive against meropenem.
Empiric antibiotic therapy for sepsis could be used meropenem [10,11]. Southwick reported that initial antibiotic, combination of ampicillin and amikacin, could be given when patient is diagnosed sepsis [11].
Discussion
Sepsis prevalence
Based on Table 1, the state could be explained because many diseases such as influenza, pneumonia and septicemia included in the top 10 of death causes in elderly. It occurred due to the reduction of immune system [12]. A number of sepsis incidences were higher on man than woman. It was related to by difference of immune responses on man and woman. Woman has good immune responses than man since woman have higher estrogen hormone. Estrogen has a role for increasing adaptive immune response. Another factor was amount of TNF (Tumor Necrosis Factor) which woman had higher than man [13].
Based on Table 2, known that almost bacteria on adult sepsis were Gram-positive bacteria. Abe et al. explained that 259 sepsis patients, 65.9% were Gram-positive and 27% were Gram-negative [14]. At the same condition at Dr. Moewardi general hospital, sepsis tended to be caused by Gram-positive. 54.71% (29 of 53 bacteria) were Gram-positive and 45.29% (24 of 53 bacteria) Gram-negative on adult sepsis patient. It can be clarified that sepsis patient with Gram-negative bacteria had IL-6 and CRP (C-Reactive Protein) higher than Gram-positive, therefore it will make a difference mechanism of PAMPs (Pathogen-Associated Molecular Patterns) [14]. IL-6 is cytokine to regulate and increase immune system against infections and tumors [15].
Resistance of gram-positive
Staphylococcus epidermidis is one of natural bacteria on skin and mucosa. But, the bacteria sometimes lead to septicemia especially people with immune system disturbance. Staphylococcus epidermidis had an ability to form biofilm [16]. Biofilm formation by bacteria will cause resistant against antibiotic. Resistance mechanism of biofilm occurred by preventing antibiotic to reach its target and decreasing efficacy of antibiotic [17]. Ziebuhr et al. reported that Staphylocoocus epidermidis, isolated from blood, had a capability to form biofilm [16]. Biofilm forming will increase bacteria resistance against antibiotic and inhibit antibiotic to reach its target [18].
Staphylococcus aureus was bacteria that cause sepsis [4]. Staphylococcus aureus was resistant against gentamicin because of antibiotic modification by acetyltransferase and phosphotransferase. On the other hand, another resistance mechanism on quinolone was the reduction of antibiotic affinity to DNA enzyme (topoisomerase IV and girase) [19]. Research on Gatermann et al. revealed that Staphylococcus epidermidis, Staphylococcus haemolyticus, and Staphylococcus hominis were the top 3 bacteria that resistant against erithromycin by percentage 62.5%, 89.8%, and 51.4%, respectively [20]. Staphylococcus hominis and Staphylococcus haemolyticus are CNS (Coagulase-Negative Staphylococci) bacteria [21]. Resistance mechanism of CNS bacteria and Staphylococcus aureus by movement of plasmid (contain resistant genes) from Staphylococci as conjugation process [22].
Resistance of gram-negative
Reduction of sensitivity on meropenem happened because of metallo-beta-lactamase. Escherichia coli that produce the enzyme will resistant 64-folds against meropenem. This was proven by raising MIC Escherichia coli with metallo-betalactamase (128 μg/mL) and without it (2 μg/mL) [23].
Resistance mechanism of Gram-negative bacilli through 3 resistance mechanisms those are producing enzyme to destroy antibiotic, mutation of antibiotic target, and efflux pump. A common mechanism on Gram-negative was beta lactamase production.
For Enterobacteriaceae groups, resistance mechanism took place by destructing antibiotic through hydrolysis and mutation of DNA gyrase and DNA topoisomerase IV. Another resistance mechanism was a modification of antibiotic. Antibiotic modification occurred on aminoglycoside by aminoglycoside acetyltransferase, aminoglycoside Onucleotidultransferase, and aminoglicoside Ophosphotransferase [24].
Generally, resistance mechanism of Pseudomonas aeruginosa against to all of antibiotic classes through reduction of cell membrane permeability and efflux pumps [25]. The usage of antibiotics, for instance penicillin, cephalosporine, and carbapenem, would induce Pseudomonas aeruginosa to produce beta lactamase [26]. On the other hand, resistance mechanisms of Acinetobacter spp. were divided into 3 categories including antibiotic inactivation by enzyme, prevention of antibiotic to reach its target, and mutation of antibiotic target. Inactivation antibiotic by Acinetobacter through producing beta lactamase and destruction of antibiotics such as penicillin, cephalosporine, and carbapenem. Some Acinetobacter strains had an ability to synthesize metallo-beta-lactamase for hydrolysis broad spectrum antibiotics included carbapenem. Another mechanism of Acinetobacter baumannii resistance was through efflux pump or aminoglycoside-modifying enzymes for aminoglycoside group [27].
Conclusion
Based on the research, we could conclude that bacteria pattern on adult sepsis patient at Dr. Moewardi general hospital was Staphylococcus haemolyticus (15.09%), Staphylococcus hominis (15.09%), Escherichia coli (13.21%), and Acinetobacter baumannii (11.32%). Resistance patterns with level of resistance more than 50% as Staphylococcus haemolyticus (ciprofloxacine, erithromycine, levofloxacine , moxifloxacine, and gentamycin ), Escherichia coli (gentamycin, ciprofloxacine, and levofloxacine), Acinetobacter baumannii (gentamycin, ciprofloxacine, levofloxacine, and ceftazidime).
11076
References
- World Health Organization (2001) Infections and infectious diseases: A manual for nurses and midwives in the WHO European Region. Europe.
- Seifert H (2009) The Clinical Importance of Microbiological Findings in the Diagnosis and Management of Bloodstream Infections. Clinical Infectious Diseases 4: 238-245.
- Cunha BA (2008) Sepsis and septic shock: selection of empiric antimicrobial therapy. Critical care clinics 24: 313-334.
- SWAB (2010) SWAB guidelines for Antibacterial Antibacterial therapy of adult patients with Sepsis. Netherland: StichtingWerkgroepAntibioticabeled.
- Artero A, Zaragoza R, Nogueira JM (2012) Epidemiology of Severe Sepsis and Septic Shock. Croatia: In Tech.
- US Department of Health and Human Services (2013) Developing Resistance. In: Antibiotic Resistance Threats in the United States 2013. Centers for Diseases Control and Prevention.
- Duerink DO, Lestari ES, Hadi U, Nagelkerke NJD, Verbrugh HA, et al. (2007) Determinants of carriage of resistant Escherichia coli in the Indonesian population inside and outside hospitals, Journal of Antimicrobial Chemotherapy 60: 377-384.
- Lewis MT, Biedenbach DJ, Jones RN (1999) In Vitro Evaluation of Cefepime and Other Broad-Spectrum beta-Lactams Against Bacteria from Indonesian Medical Centers The Indonesia Antimicrobial Resistance Study. diagnmicrobiol infect dis 8893: 285-290.
- Radji M, Fauziah S, Aribinuko N (2011) Antibiotic sensitivity pattern of bacterial pathogens in the intensive care unit of Fatmawati Hospital, Indonesia. Asian Pacific journal of tropical biomedicine 1: 39-42.
- Dipiro JT, Talbert RL, Yee GC, Matzke GR, Wells BG, et al. (2008) Pharmacotherapy A Pathophysiologic Approach. (7thedn), New York, McGraw-Hill.
- Southwick FS (2007) Infectious Disease A Clinical Short Course. (2ndedn), USA, McGraw-Hill.
- Hawkley LC, Cacioppo JT (2004) Stress and the aging immune system, Brain, Behavior, and Immunity.18: 114-119.
- Berkowitz DM, Martin GS (2010) Sepsis and Sex Can We Look Beyond Our Hormones? Chest 132: 1725-1727.
- Abe R, Oda S, Sadahiro T, Nakamura M, Hirayama Y, et al. (2010) Gram-negative bacteremia induces greater magnitude of inflammatory response than Gram-positive bacteremia. Critical Care 14: 1-7.
- Xing Z, Gauldie J, Cox G, Baumann H, Jordana M, et al. (1998) IL-6 is an antiinflammatory cytokine required for controlling local or systemic acute inflammatory responses. The Journal of clinical investigation 101: 311-320.
- Ziebuhr W, Krimmer V, Rachid S, Loßner I, Got F (1999) A novel mechanism of phase variation of virulence in Staphylococcus epidermidis?: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Molecular Microbiology 32: 345-356.
- Otto M (2008) Staphylococcal Biofilm. Curr Top MicrobiolImmunol 322: 207-228.
- Zheng Z, Stewart PS (2002) Penetration of Rifampin through Staphylococcus epidermidis Biofilms, Antimicrobial Agents and Chemotherapy 46: 900-903.
- Lowy FD (2003) Antimicrobial resistance: the example of Staphylococcus aureus. Journal of Clinical Investigation 111: 1265-1273.
- Gatermann SG, Koschinski T, Friedrich S (2007) Distribution and expression of macrolide resistance genes in coagulase-negative staphylococci. Clinical Microbiology and Infection 13: 777-781.
- John JF, Harvin AM (2007) History and evolution of antibiotic resistance in coagulase-negative staphylococci?: Susceptibility profiles of new anti-staphylococcal agents. Therapeutics and Clinical Risk Management 3: 1143-1152.
- Huebner J, Goldmann DA (1999) Coagulase-negative staphylococci: role as pathogens. Annual review of medicine 50: 223-236.
- Moloughney JG, Thomas JD, Toney JH (2005) Novel IMP-1 metallo-ß-lactamase inhibitors can reverse meropenem resistance in Escherichia coli expressing IMP-1. FEMS Microbiology Letters 243: 65–71.
- Kocsis B, Szabó D (2013) Antibiotic resistance mechanisms in Enterobacteriaceae. In: Méndez-Vilas A editor:Microbial pathogens and strategies for combating them- science, technology and education. Spain: Formatex
- Lambert PA (2002) Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. Journal of the Royal Society of Medicine 4: 22-26.
- Hancock REW, Speert DP (2000) Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resistance Updates 3: 247-255.
- Maragakis LL, Perl TM (2008) Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clinical Infectious Diseases 46: 1254-1263.








