Arthi Chandran, Samuel Kadavakollu*, Shawn White, Kiran K Andra, Naveen M Singh and Rahul Vemula
Department of Natural Sciences, Western New Mexico University, 1000 W. College Ave, Silver City, NM 88062
Corresponding Author:
Samuel Kadavakollu
Assistant Professor of Medicinal Chemistry and Shawn R. White, Ph.D.
Professor of Chemistry, Western New Mexico University
1000 W. College Ave, Silver City, NM 88062, USA
Tel: (575) 538 6641
E-mail: Samuel.Kadavakollu@wnmu.edu
Received Date: November 24, 2015; Accepted Date: December 11, 2015; Published Date: December 21, 2015
Citation: Kadavakollu S. Translational Fidelity Mediated Regulation of ER-Stress by Dph3. J Biomed Sci. 2016, 5:1. doi:10.4172/2254-609X.100019
Copyright: © 2016 Kadavakollu S. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Keywords
Translational fidelity; dph3; HAC1; Unfolded protein response; tRNA
Introduction
The immensely important genetic information carried by DNA is critically dependent on preserving the molecular structure of DNA and proper execution of transcription and translation. Unfortunately, cells are exposed to potentially damaging factors from both within and without the cell that threaten the fidelity of protein synthesis. Chemicals such as free radicals (oxygen or nitrogen oxide) and aromatic hydrocarbons or environmental agents such as X-rays and UV-light can modify DNA bases (mutations) or cause single-stranded or double-stranded DNA breaks [1-3]. This damage can cause cell death in the extreme cases, or worse, lead to carcinogenesis [4]. Fortunately, cells have developed DNA damage response mechanisms which regulate gene expression to coordinate proper DNA repair and replication [5].
In addition, various biological checkpoints in the cell exist to ensure that proteins are synthesized properly from the DNA code. Cells respond to stress at the translational and post translational levels [6]. Transcriptional regulation of gene expression is the main mode of regulation of gene expression; however, for certain genes, translational regulation also plays a major role. During the step of translational elongation, the tRNA, which is an adaptor molecule between the mRNA and the protein, serves an important role in the translational regulation of cellular responses to stress and is a biomolecules which is very heavily modified [7,8]. One example of the 90+ known modified versions of nucleosides that exist are uracil modifications which have been shown to improve the translation of transcripts rich in AGA and GAA codons [9-11]. These findings suggest that the anticodons containing mcm5U, mcm5s2U, and ncm5U (Table 1) modifications at the wobble position have enhanced affinity for specific codons and such modifications can regulate the speed of translation elongation [8]. Thus, in a mixed box, these modifications can enhance binding to A-ending codons and restrict binding to U- and C- ending codons [9-11]. A failure to restrict anticodon binding to U- and C- ending codons leads to incorporation of the wrong amino acid and promotes translational infidelity.
ψ
| Codon |
Anti codon |
Amino
acid |
Codon |
Anti codon |
Amino
acid |
Codon |
Anti codon |
Amino
acid |
Codon |
Anti codon |
Amino
acid |
| UUU |
AAA |
Phe |
UCU |
AGA |
Ser |
UAU |
AUA |
Tyr |
UGU |
ACA |
Cys |
| UUC |
GAA |
UCC |
GGA |
UAC |
GΨA |
GUC |
GCA |
| UUA |
ncm5UmAA |
Leu |
UCA |
ncm5UGA |
UAA |
UUA |
Stop |
UGA |
UCA |
Stop |
| UUG |
m5CAA |
UCG |
CGA |
UAG |
CUA |
UGG |
CCA |
Trp |
| CUU |
AAG |
Leu |
CCU |
AGG |
Pro |
CAU |
AUG |
His |
CGU |
ACG |
Arg |
| CUC |
GAG |
CCC |
GGG |
CAC |
GUG |
CGC |
GCG |
| CUA |
UAG |
CCA |
ncm5UGG |
CAA |
mcm5s2UUG |
Gln |
CGA |
UCG |
| CUG |
CAG |
CCG |
UGG |
CAG |
CUG |
CGG |
CCG |
| AUU |
AAU |
lle |
ACU |
AGU |
Thr |
AAU |
AUU |
As n |
AGU |
ACU |
Ser |
| AUC |
GAU |
ACC |
GGU |
AAC |
GUU |
AGC |
GCU |
| AUA |
ΨAΨ |
ACA |
ncm5UGU |
AAA |
mcm5s2UUU |
Lys |
AGA |
mcm5UCU |
Arg |
| AUG |
CAU |
Met |
ACG |
CGU |
AAG |
CUU |
AGG |
CCU |
| GUU |
IAU |
Val |
GCU |
AGC |
Ala |
GAU |
AUC |
As p |
GGU |
ACC |
Gly |
| GUC |
GAC |
GCC |
GGC |
GAC |
GUC |
GGC |
GCC |
| GUA |
ncm5UAC |
GCA |
ncm5UGC |
GAA |
mcm5s2UUC |
Glu |
GGA |
mcm5UCU |
| GUG |
CAC |
GCG |
CGC |
GAG |
CUC |
GGG |
CCC |
Table 1: Genetic code including modified bases. Mixed codon boxes are shown in grey. Pseudouridine is shown as the Greek letter Ψ.
Dph3 (also known as Kti11) is one of the five proteins (Dph1 to Dph5) required for diphthamide biosynthesis [12] although it also has a known, second role in the effective functioning of the Elongator complex (Elp1 to Elp6) [13,14]. Recently, Nedialkova and Leidel examined the role of uracil modifications in optimal codon translation rates and showed the formation of protein aggregates in ncs2Δelp6Δyeast [15]. Since Dph3 is also known to be involved in the wobble uracil modification [16-18], we predicted that a deficiency in Dph3-catalyzed modifications would allow binding of anticodons to near-cognate codons and promote translational infidelity. Pharmaceutical agents that promote translational infidelity can be used to phenocopy the wobble uracil modification deficiency [19]. Aminoglycoside antibiotics, such as paromomycin, target the region of codon-anticodon pairing and cause translational errors by altering the dynamics of codon-anticodon interactions. While misincorporation at premature stop codons can be used to rescue proteins from premature termination of translation, it also has the potential to promote errors and lead to misfolded proteins [20]. Misfolded proteins are, in general, insoluble, easily aggregate and are toxic to cells. In order to prevent toxicity, these proteins are refolded in the endoplasmic reticulum. As the cell accumulates misfolded proteins, it triggers a cascade of pathways called the endoplasmic reticulum stress response or ER-stress pathway. ER-stress is involved in the quality control of proteins in the endoplasmic reticulum (ER) and targets proteins that cannot be successfully folded for degradation via an endoplasmic reticulum assisted degradation pathway [21,22].
The unfolded proteins in the ER are folded by the ER-chaperone called as Kar2. In the process of folding, the Kar2 dissociates from Ire1, leading to dimerization and activation of Ire1. The activated Ire1 splices HAC1 mRNA in the cytoplasm, resulting in the translation of HAC1 mRNA into Hac1 protein. The Hac1 protein enters the nucleus and binds to UPRE (unfolded protein response element) which promotes genes involved in protein folding including HAC1 [23,24]. Thus the synthesis of proteins involved in protein folding is amplified by hyperactivation of ER-stress, ultimately leading to increased protein folding. The paromomycin binds at the codon-anticodon interface in the ribosome, increasing translational errors and leading to increased production of unfolded proteins. Aminoglycoside antibiotics have been previously shown [9] to increase splicing of HAC1 and transcription of Kar2 indicating that translational errors can indeed hyperactivate ER-stress.
In this paper, the role of Dph3 in the prevention of translational errors was examined. Cytotoxicity studies were performed with paromomycin on wild-type, dph3 mutants, and dph3 mutants with the re-expressed DPH3 gene. In addition, UPR reporters such as HAC1 and lacZ were used to investigate more closely the action of Dph3 in the cascade of translational events.
Materials and Methods
Transformation
The wild type and dph3 mutants were transformed with control vector BY011 (galactose-inducible, ampicillin resistance in E. coli., URA3 resistance in yeast) and pre-cloned DPH3 under GAL1 promoter in BY011 (available in HIP FLEXGene Saccharomyces cerevisiae ORF collection ) as described in the paper [25]. The URA 3 marker from pYES2.0 plasmid was PCR amplified using the following primers: Forward: 5’-GAGATCCAGTTCGATGTAACC-3’ and Reverse: 5’-ATCACACTGCCTTTGCTGAG-3’. Then, the cassette for deletion of DPH3 was generated by PCR amplification using the following primers: Forward: 5’-ATGTCAACATATGACGAAATCGAAATCGAAGAGATCCAGTTCGATGTAACC- 3’ and Reverse: 5’-TTAGGCAGCAGCGG-CAATAGGCTCAGGGGGATCACACTGCCTTT GCTGAG-3’. The deletion cassette was transformed into BY4741 strains using EZ yeast transformation kit (Zymo Research, Irvine, CA) and the transformants were selected on SD-Ura plates. The MET15 marker from pRS401 plasmid was PCR amplified using the following primers: Forward: 5’-CTGTGCGGTATTTCACACCG-3’ and Reverse: 5’-AGATTGTACTGAGAGTGCAC-3’. Then, the cassette for deletion of DPH3 was generated by PCR amplification using the following primers: Forward: 5’-ATGTCAAC-ATATGACGAAATCGAAA TCGACTGTGCGGTATTTCACACCG-3’ and Reverse: 5’-TTAGG-CAGCA GCGGCAATAGGCTCAGGGGGAGATTGTACTGAGAGTGCAC-3’. The deletion cassette was transformed into AWY14 [26] strains using EZ yeast transformation kit (Zymo Research, Irvine, CA) and the transformants were selected on SD-Met plates. Finally, gene deletion of DPH3 in BY4741 and AWY14 strains was confirmed using the primers: Forward 5’-AGTTCCATCCCGTAACACCA-3’ and Reverse: 5’-TGTCACAGCCATTTGAGATGA-3’.
Cytotoxicity Assay
For sensitivity assays, the overnight culture of wild-type, dph3 mutants, and dph3 mutants with the re-expressed DPH3 gene were grown overnight in SD-Ura media and plated in SGal/-Ura plates containing 100 μg/ml of paromomycin. The plates were incubated at 30°C for 3 days and imaged using BIO Rad Gel Doc EZ imaging system.
Unfolded Protein Response Marker Study
AWY14+BY011, AWY14+BY011-dph3, AWY14+BY011- dph3+DPH3, AWY14+ BY011+ paromomycin, AWY14+BY011- dph3+paromomycin and AWY14+BY011-dph3+DPH3+ paromomycin were grown overnight in SD-Ura, Cells were taken at 104 cells/ml, grown to 5x106 cells/ml and then induced for 2 hours in SGal-Ura media containing 100 μg/ ml of paromomycin at 30°C. The cells were spun down, the protein extracted, and the lacZ assay performed using lacZ assay kit (Thermofisher).
Northern blots
Both the dph3 mutant and wild-type cells were grown to ˜5 × 106 cells/ml in YPD and treated with 100 μg/ml paromomycin for 60 minutes. Afterwards, the RNA was carefully purified using the RNeasy Mini Kit and analyzed as described [10]. Detection of the RNA was facilitated using the Chemiluminescent Nucleic Acid Detection Module [10] (Pierce; Rockford, IL).
Results
dph3 mutants have a higher level of translational infidelity. The AWY14 wild type and dph3 mutants cells that were grown overnight in SD-ura were stressed using paromomycinan aminoglycoside antibiotic known to induce translational errors. The dph3 mutants showed an increased sensitivity to paromomycin (Figure 1) when observed after incubation at 30°C for three days. As seen in Figure 1, these mutants can be rescued from paromomycin by overexpressing DPH3 in the mutants. Furthermore, the full effect of the cytotoxic agent is readily apparent in the mutants at 100 μg/mL, with large increase in dose having little discernible effect.
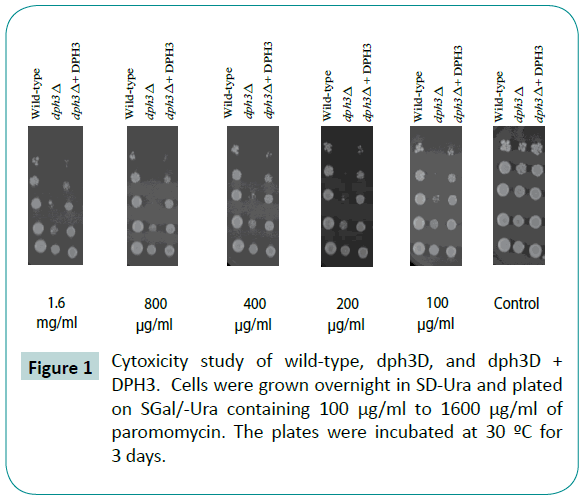
Figure 1: Cytoxicity study of wild-type, dph3D, and dph3D + DPH3. Cells were grown overnight in SD-Ura and plated on SGal/-Ura containing 100 μg/ml to 1600 μg/ml of paromomycin. The plates were incubated at 30 ºC for 3 days.
The dph3 mutants have an increased unfolded protein response reporter activity under basal conditions, which is further increased in the presence of paromomycin.
AWY14 cells were used where the lacZ gene is placed under the control of KAR2 promoter containing the UPRE element. The binding of Hac1 protein to UPRE element initiates the transcription of lacZ and KAR2 genes proportionately. The amount of lacZ protein levels in the cells is a direct indicator of Kar2 protein levels, and thus a measure of amount of unfolded proteins present in the cells. This conclusion is based on previous work [27] where the lacZ gene was put under the control of UPRE (unfolded protein response element). These researchers showed that since UPRE mainly regulates transcription of KAR2, the KAR2 transcript levels should be proportional to lacZ levels.
The AWY14-dph3 mutants have an increased UPR reporter activity (Figure 2) under basal conditions compared to wild-type AWY14 cells with the UPR reporter activity further increased in the presence of paromomycin.
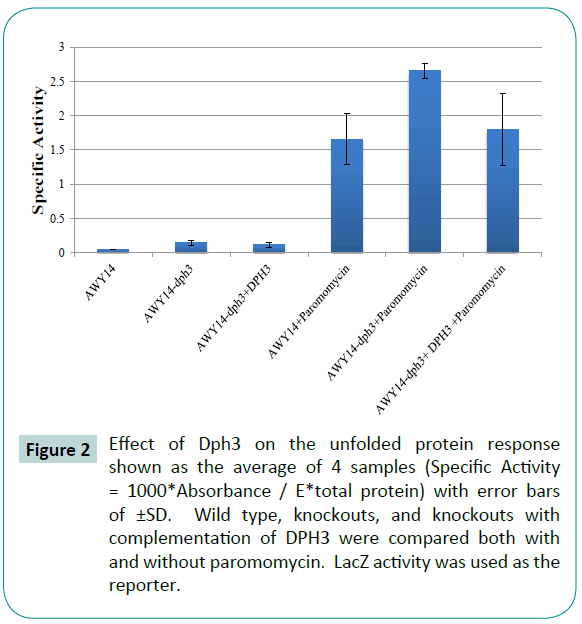
Figure 2: Effect of Dph3 on the unfolded protein response shown as the average of 4 samples (Specific Activity = 1000*Absorbance / E*total protein) with error bars of ±SD. Wild type, knockouts, and knockouts with complementation of DPH3 were compared both with and without paromomycin. LacZ activity was used as the reporter.
The dph3 mutants have increased HAC1 mRNA splicing.
The northern blots of wild-type and dph3 mutants were performed after cells were grown to mid log phase. One group was measured under basal conditions and the other treated with paromomycin for 1 hour. Figure 3 shows increased HAC1 splicing in dph3 mutants.
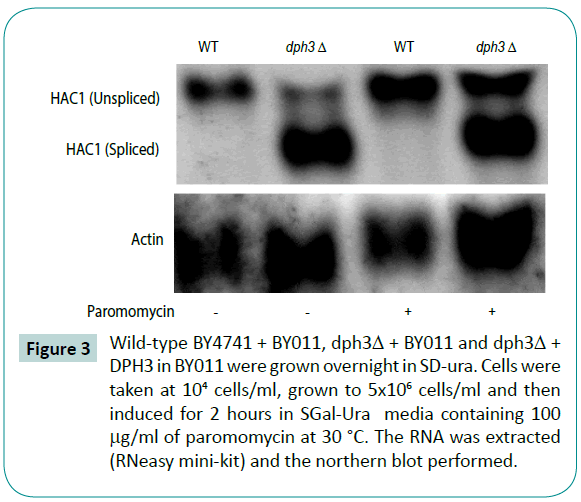
Figure 3: Wild-type BY4741 + BY011, dph3Δ + BY011 and dph3Δ + DPH3 in BY011 were grown overnight in SD-ura. Cells were taken at 104 cells/ml, grown to 5x106 cells/ml and then induced for 2 hours in SGal-Ura media containing 100 μg/ml of paromomycin at 30 °C. The RNA was extracted (RNeasy mini-kit) and the northern blot performed.
Discussion
The dph3 mutants have a heightened sensitivity to paromomycin which is a known translational error inducer (Figure 1). Since Dph3 is involved in the modification of the wobble uracil of the tRNAs [28] in the mixed codon boxes, the codon-anticodon interactions (Table 1) in dph3 mutants might be perturbed, giving rise to translational errors like aminoacid misincorporation, frameshifting and premature termination of translation. The translational errors would then give rise to unfolded proteins which must be folded by chaperones like Kar2 in endoplasmic reticulum. During the process of folding, Kar2 also leads to activation of Ire1, a transmembrane sensor in ER membrane. The activated Ire1 in turn causes the splicing of HAC1 mRNA, leading to the translation of spliced HAC1 mRNA into Hac1 protein. The increased splicing of HAC1 mRNA in paromomycin treated dph3 mutants observed therefore, indicates the increased basal rate of ER-stress activation in these mutants (Figure 3). Also, the HAC1 splicing also occurs in wild-type cells upon paromomycin treatment, but not as much as compared to dph3 mutants, confirming translational errors indeed cause increased splicing of HAC1 mRNA.
The Hac1 protein synthesized by the translation of spliced HAC1 mRNA enters the nucleus and binds to UPRE elements in protomers of genes like KAR2, causing their increased transcription and thus increasing the synthesis of Kar2 protein in response to unfolded proteins. The lacZ reporter used indicates the rate of transcription of genes regulated by UPRE like Kar2 and was observed to increase the activity of the UPR reporter in dph3 mutants. The reporter activity was further enhanced upon treatment with paromomycin indicating increased levels of unfolded proteins in dph3 mutants (Figure 2). Since the unfolded protein levels measured by UPR reporter and HAC1 splicing increased after paromomycin treatment we can conclude that it is indeed the increased translational errors in dph3 mutants that gives rise to hyperactivation of ER stress in the mutants.
Conclusion
This study revealed the critical role of Dph3 in the cell’s ability to produce proteins accurately. dph3 mutants showed a lowered viability especially when under chemical stress. In addition, the heightened levels of unfolded protein response markers observed in mutants lacking DPH3, demonstrated that this gene in Saccharomyces cerevisiae was indeed involved in the prevention of translational errors. Higher levels of lacZ in dph3 mutants were observed indicating an activated UPR. Dph3 protein added to the mutant cells achieved a partial rescue from translational errors indicated by slightly decreased lacZ levels. In addition, northern blot analysis showed an increased presence of the important UPR signaling pathway HAC1 spliced RNA in dph3 mutants.
7868
References
- Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366-374.
- Garinis GA (2008) DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol 10: 1241-1247.
- Wang JS, Engle, Y Zhang(2011) A new in vitro system for activating the cell cycle checkpoint. Cell Cycle 10: 500-6.
- Kidane D, Chae WJ, Czochor J, Eckert KA, Glazer PM, et al. (2014) Interplay between DNA repair and inflammation, and the link to cancer. Crit Rev BiochemMolBiol 49: 116-139.
- Jin BK, D Robertson (2013) DNA Methyltransferases (DNMTs), DNA Damage Repair, and Cancer. Advexper med biol 754: 3-29.
- Schafer S, Adami E, Heinig M, Rodrigues KE, Kreuchwig F, et al. (2015) Translational regulation shapes the molecular landscape of complex disease phenotypes. Nat Commun 6: 7200.
- Hori H (2014) Methylated nucleosides in tRNA and tRNAmethyltransferases. Front Genet5: 144.
- Towns WL, TJ Begley(2012) Transfer RNA Methytransferases and Their Corresponding Modifications in Budding Yeast and Humans: Activities, Predications, and Potential Roles in Human Health. DNA Cell Biol 31: 434-454.
- Patil A(2012) Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol 9: 990-1001.
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, et al. (2007) Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell 28: 860-870.
- Agris PF (2004) Decoding the genome: a modified view. Nucleic Acids Res 32: 223-238.
- Liu S, SH Leppla(2003) Retroviral insertional mutagenesis identifies a small protein required for synthesis of diphthamide, the target of bacterial ADP-ribosylating toxins. Mol Cell 12: 603-613.
- Fichtner L (2003)Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. MolMicrobiol, 49: 1297-1307.
- Jablonowski D, Frohloff F, Fichtner L, Stark MJ, Schaffrath R (2001) Kluyveromyceslactiszymocin mode of action is linked to RNA polymerase II function via Elongator. MolMicrobiol 42: 1095-1105.
- Nedialkova DD, Leidel SA (2015) Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 161: 1606-1618.
- Bar C (2008)A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes: Kti11/Dph3. MolMicrobiol69: 1221-1233.
- Glatt S, Zabel R, Vonkova I, Kumar A, Netz DJ, et al. (2015) Structure of the Kti11/Kti13 heterodimer and its double role in modifications of tRNA and eukaryotic elongation factor 2. Structure 23: 149-160.
- Zabel R(2008) Yeast alpha-tubulin suppressor Ats1/Kti13 relates to the Elongator complex and interacts with Elongator partner protein Kti11. MolMicrobiol69: 175-187.
- Kramer EB (2010) A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA16: 1797-1808.
- Paredes JA, Carreto L, Simões J, Bezerra AR, Gomes AC, et al. (2012) Low level genome mistranslations deregulate the transcriptome and translatome and generate proteotoxic stress in yeast. BMC Biol 10: 55.
- Verghese J2012) Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. MicrobiolMolBiol Rev76: 115-158.
- Wu H, Ng BS, Thibault G (2014) Endoplasmic reticulum stress response in yeast and humans. Biosci Rep 34.
- Herzog B, (2013) Mutual cross talk between the regulators Hac1 of the unfolded protein response and Gcn4 of the general amino acid control of Saccharomyces cerevisiae. Eukaryot Cell12: 1142-1154.
- Anshu A (2015) A novel role for protein kinase Kin2 in regulating HAC1 mRNA translocation, splicing, and translation. Mol Cell Biol 35: 199-210.
- Fleming MS, Gitler AD (2011) High-throughput yeast plasmid overexpression screen. J Vis Exp .
- Liu Y (2000) Characterization of a Saccharomyces cerevisiae homologue of Schizosaccharomycespombe Chk1 involved in DNA-damage-induced M-phase arrest. Mol Gen Genet 262: 1132-1146.
- Schröder M, Clark R, Kaufman RJ (2003) IRE1- and HAC1-independent transcriptional regulation in the unfolded protein response of yeast. Mol Microbiol 49: 591-606.
- Liu S, Wiggins JF, Sreenath T, Kulkarni AB, Ward JM, et al. (2006) Dph3, a small protein required for diphthamide biosynthesis, is essential in mouse development. Mol Cell Biol 26: 3835-3841.