B Sharan Sharma1,2*
1 Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan
2 Department of Life Sciences, Indrashil University, Kadi, Gujarat, India
- *Corresponding Author:
- Sharma BS
Department of Life Sciences
Indrashil University, Kadi, Gujarat, India.
Tel: +91-2764278813
E-mail: bssgenetix@gmail.com
Received Date: May 09, 2018; Accepted Date: June 04, 2018; Published Date: June 11, 2018
Citation: Sharma BS (2018) Ultraviolet- Induced DNA Damage Promotes Methylation of the Cell-Cycle Checkpoint Kinase CHK1. Arch Cancer Res. Vol.6 No.2:10 DOI: 10.21767/2254-6081.100176
Background: Checkpoint kinase 1 (CHK1) is a serine/threonine-protein kinase which plays a major role during checkpoint-mediated cell cycle arrest and activation of DNA repair in response to DNA damage. Ultraviolet (UV) irradiation induces DNA damage triggering the cell cycle arrest and the cell may be forced into apoptosis if damage is not repaired. Through phosphorylation mediated activation CHK1, as an effector kinase, responds to this damage by targeting downstream effector proteins.
Objective: The objective of the present study was to demonstrate CHK1 methylation in response to UV induced DNA damage.
Methods: Expression of CHK1 was detected in a cancer cell line Hct-116. Hct-116 cells, cultured in RPMI-1640 medium supplemented with serum and antibiotics, were transfected with HA-Chk1 plasmid, exposed to UV radiation and incubated for different time intervals. Cell lysis followed by immunoblotting was performed to visualize the signals of methylated CHK1.
Result: Methylated CHK1 signals were observed in response to UV induced DNA damage in a cancer cell line Hct-116. Expectedly, it was found that with the increased duration of post UV exposure, methylation level of CHK1 was also amplified.
Discussion and Conclusion: Here, for the first time, it is reported that DNA damage induced by UV radiation was associated with elevated methylation of CHK1. This new finding might indicate that cells may have evolved mechanisms to promote CHK1 methylation for reasons not yet known. This study reveals novel modification of CHK1 as a component of the cellular response to DNA damage which may help us understand the importance of such modifications.
Keywords
CHK1 methylation; Hct-116 cell line; Immunoblotting; UV induced DNA damage
Introduction
To counter DNA damage, cells initiate repair mechanism by means of cell cycle checkpoints. Checkpoint kinase 1 (CHK1), as a key regulator of checkpoint signalling, is one of the vital protein kinases of this repair pathway. CHK1 has an important role in normal cell cycle checkpoints, cell proliferation, and viability in all eukaryotes [1,2]. In response to DNA damage, CHK1 is phosphorylated and activated by ataxia telangiectasia and Rad3 related (ATR) [1,3]. CHK1 is composed of a highly conserved kinase domain at the amino (N)-terminal half and a regulatory region at the carboxyl (C)-terminal half, and primarily responds to replication fork interference in the S phase and DNA damage at the G2 phase [4,5]. Exposure to ultra-violet (UV) irradiation triggers the formation of DNA lesions in the cells in the form of DNA intra-strand cross-links in genomic DNA [6]. These lesions can be both cytotoxic and mutagenic and can trigger cell death or accelerated ageing and can lead to the onset of skin cancer [7]. The induction of DNA damage by UV irradiation triggers the dissociation of CHK1 from chromatin, followed by the de phosphorylation of H3T11P by phosphatase PP1g, which results in the DNA damage-induced repression of genes such as cyclin B1 and Cdk1 [8,9]. DNA methylation is a vital epigenetic modification necessary for normal regulation of genes which gets frequently decontrolled in cancer [10]. The discovery of lysine and arginine methylation in histones and other proteins and the enzymes that carry out these posttranslational modifications has added a new dimension to the signal transduction field [11]. Phosphorylation of CHK1 at two conserved ATR sites, Ser-317 and Ser-345, has now been known for the activation of replication checkpoints. However, CHK1 methylation has never been studied in response to DNA damage. In this study, it was uncovered that exposure of UV irradiation leads to elevated methylation of CHK1 as a component of the cellular response to DNA damage. Such modification, discovery of Methylated form of CHK1 in response to DNA damage after UV treatment, is thought provoking and will lead to further studies to understand the precise mechanism of CHK1 methylation.
Materials and Methods
Cell culture and treatments
Hct-116 cells were maintained in RPMI-1640 media supplemented with 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin. Hct-116 cells were transfected with HA-CHK1 plasmid, transfection was performed by calcium phosphate precipitation method. The construct was kindly provided by Dr. Sheau-Yann Shieh, IBMS, Academia Sinica. The cells were exposed to 20 J/m2 UV irradiation and incubated for different time intervals (0, 0.5, 1 and 2 hrs) for DNA damage experiments.
Cell lysis and immunoblotting
To examine the methylation level of CHK1 after DNA damage, cell lysates were prepared in TEGN buffer (10 mM Tris at pH 7.5, 1 mM EDTA, 420 mM NaCl, 10% glycerol, and 0.5% Nonidet P-40) containing 10 mM NaF, 10 mM glycerophosphate, 1 mM sodium orthovanadate, 1 mM DTT, and protease inhibitor mixture (Roche). The antibodies used for immunoblotting were as follows: rabbit anti-actin (Sigma), mouse anti-methylated lysine (Sigma), and mouse anti-HA (Sigma). Anti-mouse HRP (Roche) and antirabbit HRP (Roche) were used as secondary antibody. The bands were resolved by 8-10% SDS/PAGE followed by western blot. The membrane was probed with either methylation specific (antimethylated lysine) antibody or CHK1 specific (anti-HA) antibody and signals were detected.
Result
CHK1 was methylated in response to UV induced DNA damage
To evaluate whether CHK1 can be methylated in response to DNA damage, first the blot was probed with methylation specific antibody (anti-methylated lysine) and signals of methylated lysine (Met Lysine) were detected. It was found that in response to UV induced DNA damage methylated CHK1 signals (~55kDa) could be observed (recognized by Met Lysine - proteins methylated on lysine residues) (Figure 1). CHK1 methylation level was nil in the 0hr treatment but as the duration of incubation increased post UV treatment, methylation level of CHK1 was also increased (Figure 1). Middle panel in the Figure 1 is showing the presence of CHK1 after having stripped the blot and re-probed it with anti- HA antibody.
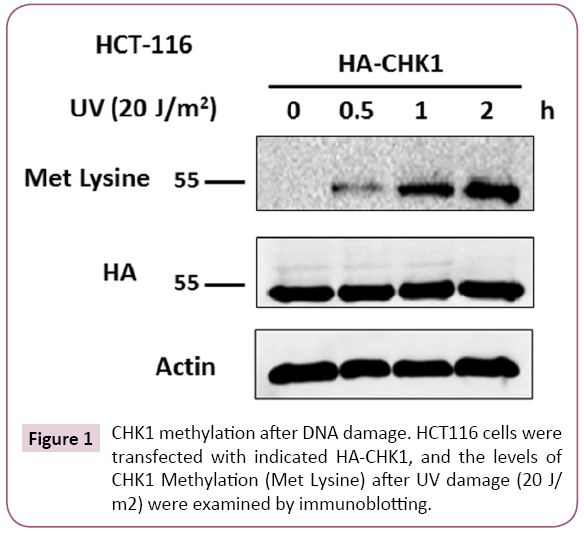
Figure 1: CHK1 methylation after DNA damage. HCT116 cells were transfected with indicated HA-CHK1, and the levels of CHK1 Methylation (Met Lysine) after UV damage (20 J/ m2) were examined by immunoblotting.
Discussion and Conclusion
In this study, methylated form of CHK1 was identified. For the first time, it was discovered that CHK1 is methylated in response to UV induced DNA damage. Most importantly, CHK1 is a vital checkpoint protein which plays a major role during DNA damage. DNA methylation, as a vital epigenetic modification, is involved at many levels of gene regulation and signal transduction [11]. Post translational modifications of CHK1 are known for example Cheng et al., [12] demonstrated the functional interplay among BTG3, CHK1, and CRL4Cdt2 and suggested that genotoxic stress promotes the interaction and K63-linked ubiquitination of CHK1. In another study by the same author, it was demonstrated that BTG3 overexpression and CHK1 methylation are linked [13]. CHK1 has been shown to be active even in unperturbed cell cycles [14,15], and although it is further activated in response to DNA damage or stalled replication, this may not require CHK1 dimerization or auto-phosphorylation [16]. Present study explored the methylation status of CHK1 in response to genotoxic stress and increased methylation of CHK1 was observed. This discovery indicates that cells may have evolved mechanisms to promote CHK1 methylation for reasons not yet known. Many protein kinases possess an integral regulatory domain that mediates a variety of functions including interactions with other proteins, activation, or inhibition of the kinase domain [13,17]. One of the limitations of this study was not exploring the methylation level of CHK1 mutants such as Kinase dead or N-terminal deleted mutants in response to genotoxic stress which leads to an interesting question ‘what region/domain of CHK1 was methylated?’ In a previous work, it was observed that only wild type CHK1, but not the CHK1 kinase domain impaired mutants, was modified for methylation when co-expressed with candidate tumor suppressor BTG3 gene, suggesting the important role of kinase domain for such modification [13]. Another question is what are the important residues of CHK1 which sit at the key interface between the N-terminal kinase domain and the C-terminal domain and if one of them is mutated then does it lead to methylation of CHK1 in the absence of DNA damage? Clearly, in-depth structural studies are needed to respond to these questions. The exploration of CHK1 methylation, as described in this study, may lead to further studies to understand methylated CHK1-regulated signal transduction.
Conflicts of Interest
The author has no conflicts of interest to disclose.
Acknowledgments
The author thanks Dr. Sheau-Yann Shieh for providing the materials and lab space for conducting the research. Dr. Yu-Che Cheng is thankfully acknowledged for his guidance.
22738
References
- Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, et al. (2000) Chk1 is an essential kinase thatisregulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev 14:1448-1459.
- Zaugg K, Su YW, Reilly PT, Moolani Y, Cheung CC, et al. (2007) Cross-talk between Chk1 and Chk2 in double-mutant thymocytes. Proc NatlAcadSci104:3805-3810.
- Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, et al. (2000) Chk1 is an essential kinase thatisregulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev 14:1448-1459.
- Zhao HU, Piwnica WH (2001) ATR-mediated checkpoint pathwaysregulate phosphorylation and activation of human Chk1. Mol Cell Biol21:4129-4139.
- Zaugg K, Su YW, Reilly PT, Moolani Y, Cheung CC, et al. (2007) Cross-talk between Chk1 and Chk2 in double-mutant thymocytes. Proc NatlAcadSci104:3805-3810.
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, et al. (1997) Mitotic and G2 checkpoint control:Regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277:1501-1505.
- Zhao HU, Piwnica WH (2001) ATR-mediated checkpoint pathwaysregulate phosphorylation and activation of human Chk1. Mol Cell Biol21:4129-4139.
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, et al. (1997) Conservation of the Chk1 checkpoint pathway in mammals:Linkage of DNA damage to Cdkregulationthrough Cdc25. Science 277:1497-1501.
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, et al. (1997) Mitotic and G2 checkpoint control:Regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277:1501-1505.
- Jackson SP, Bartek J (2009) The DNA-damage response in humanbiology and disease. Nature 461:1071-1078.
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, et al. (1997) Conservation of the Chk1 checkpoint pathway in mammals:Linkage of DNA damage to Cdkregulationthrough Cdc25. Science 277:1497-1501.
- Mitchell JR, Hoeijmakers JH, Niedernhofer LJ (2003) Divide and conquer:Nucleotide excision repair battles cancer and ageing.CurrOpinCell Biol15:232-240.
- Jackson SP, Bartek J (2009) The DNA-damage response in humanbiology and disease. Nature 461:1071-1078.
- Shimada M, Haruta M, Niida H, Sawamoto K, Nakanishi M (2010) Protein phosphatase 1γ isresponsible for dephosphorylation of histone H3 at Thr 11 after DNA damage. EMBO Rep 11: 883-889.
- Mitchell JR, Hoeijmakers JH, Niedernhofer LJ (2003) Divide and conquer:Nucleotide excision repair battles cancer and ageing.CurrOpinCell Biol15:232-240.
- Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, et al. (2008) Chk1 is a histone H3 threonine 11 kinase thatregulates DNA damage-inducedtranscriptionalrepression. Cell132:221-232.
- Shimada M, Haruta M, Niida H, Sawamoto K, Nakanishi M (2010) Protein phosphatase 1γ isresponsible for dephosphorylation of histone H3 at Thr 11 after DNA damage. EMBO Rep 11: 883-889.
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev16:6-21.
- Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, et al. (2008) Chk1 is a histone H3 threonine 11 kinase thatregulates DNA damage-inducedtranscriptionalrepression. Cell132:221-232.
- Lee DY, Teyssier C, Strahl BD, Stallcup MR (2005) Role of proteinmethylation in regulation of transcription. EndocrRev26:147-170.
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev16:6-21.
- Cheng YC, Lin TY, Shieh SY (2013) Candidate tumorsuppressor BTG3 maintainsgenomicstability by promoting Lys63-linked ubiquitination and activation of the checkpoint kinase CHK1. Proc NatlAcadSci110:5993-5998.
- Lee DY, Teyssier C, Strahl BD, Stallcup MR (2005) Role of proteinmethylation in regulation of transcription. EndocrRev26:147-170.
- Sharma BS (2017) BTG3, a candidate tumorsuppressor, promotesmethylation of checkpoint kinase CHK1. Gene Reports 7:119-122.
- Cheng YC, Lin TY, Shieh SY (2013) Candidate tumorsuppressor BTG3 maintainsgenomicstability by promoting Lys63-linked ubiquitination and activation of the checkpoint kinase CHK1. Proc NatlAcadSci110:5993-5998.
- Zhao H, Watkins JL, Piwnica-Worms H (2002) Disruption of the checkpoint kinase 1/cell division cycle 25A pathwayabrogatesionizing radiation-induced S and G2 checkpoints. Proc NatlAcadSci99:14795-14800.
- Sharma BS (2017) BTG3, a candidate tumorsuppressor, promotesmethylation of checkpoint kinase CHK1. Gene Reports 7:119-122.
- Sørensen CS, Syljuåsen RG, Falck J, Schroeder T, Rönnstrand L, et al. (2003) Chk1 regulates the S phase checkpoint by coupling the physiologicalturnover and ionizing radiation-inducedacceleratedproteolysis of Cdc25A. Cancer Cell3:247-258.
- Zhao H, Watkins JL, Piwnica-Worms H (2002) Disruption of the checkpoint kinase 1/cell division cycle 25A pathwayabrogatesionizing radiation-induced S and G2 checkpoints. Proc NatlAcadSci99:14795-14800.
- Bartek J, Lukas J (2003) Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell3:421-429.
- Sørensen CS, Syljuåsen RG, Falck J, Schroeder T, Rönnstrand L, et al. (2003) Chk1 regulates the S phase checkpoint by coupling the physiologicalturnover and ionizing radiation-inducedacceleratedproteolysis of Cdc25A. Cancer Cell3:247-258.
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the humangenome. Science 298:1912-1934.
- Bartek J, Lukas J (2003) Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell3:421-429.
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the humangenome. Science 298:1912-1934.