Keywords
Clinical microbiology; Urinary tract infection; Quantitative urine culture; Dipslide; Plate culture; External quality assurance; EQA
Introduction
Quantitative culture of urine forms the basis for the microbiological diagnosis of urinary tract infection. This microbiological investigation is expected to pinpoint any causative agent of infection. In clinically dubious cases, it may also help to differentiate between real infection and harmless colonisation of urinary tract or just contamination of the urine specimen.
The traditional diagnostic criterion for uropathogen growth has been >105 CFU/ml, but a lower limit of 103 has been proposed for symptomatic persons [1]. Information about the patient’s symptoms rarely reaches the laboratory and, therefore, the decision on the significance of the finding to be reported to the physician has to be made without this information. The laboratory diagnosis is most often based only on the number of microbes, which increases the relevance of the correct quantitative culture of urine. Following the updating of the guidelines on urine culture in Finland [2], a questionnaire was submitted to laboratories in 2001. The results showed a tendency to lower the limit of significant growth from CFU/ml of >105 to 104 [3].
Since urinary tract infections are among the most common infections treated in health care facilities, the quantitative culture of urine is the most common clinical microbiological investigation performed in clinical laboratories. In Finland nearly 400 laboratories cultivate annually over 1.5 million urine specimens. Ensuring an acceptable level of performance of urine culture in these laboratories is not easy. The Finnish Communicable Disease Act [4] stipulates that a laboratory must have a licence to perform and report on their clinical microbiology investigations. To obtain the licence, a laboratory has to take part in External Quality Assurance (EQA), including that of quantitative urine culture. This offers a possibility to evaluate the quality of performance in urinary culture for common microbial pathogens. A valid method to monitor how well laboratories are able to perform the quantitative culture of urine specimens is to follow their success in EQA. In this study, we analysed almost 7000 EQA results of 335 laboratories participating in EQA surveys during the period of 2009- 2011.
Material and Methods
Background data on stipulations
All laboratories that participated in this study were licensed and approved by the Regional State Administrative Agencies (RSAAs) in accordance with the Finnish Communicable Disease Act [4]. The National Institute for Health and Welfare (THL) has given detailed instructions for the licensing process. The basic requirements include that each laboratory has appropriate and sufficient equipment as well as professional staff in relation to the function and test variability of the laboratory. In addition, the laboratory has to participate annually in at least four EQA rounds per type of clinical microbiological investigation. The aim of this licensing is to assure comparable and reliable laboratory performance in all licensed laboratories. By request, all laboratories have to give RSAAs and THL all relevant information related to their microbiological activities. This information includes all data on their EQA results.
Participants
The total number of the participating laboratories during the study period of three years (2009-2011) was 335. However, the number of participants in each of the four annual EQA rounds varied from 293 to 303 in 2009, from 282 to 299 in 2010 and from 273 to 283 in 2011. Of the participants, 312 were small laboratories inside health centres. Twenty-three laboratories were large, specialised clinical microbiology laboratories. All laboratories quantified the microbial growth detected in urine specimens. If the quantity of growth reached the level regarded as clinically significant, indicating infection, the growth was identified and a susceptibility test was performed. The number of laboratories performing the identification of the most common uropathogen Escherichia coli varied from 54 to 65, but 26 laboratories identified all bacteria they found in urine specimens. Most of the smaller laboratories sent the bacterial culture to the larger laboratories for identification and susceptibility testing.
EQA schemes for urine culture and reporting
In Finland, the EQA schemes are commonly organized by Labquality Ltd. The specimens for each urine EQA scheme are designed by a Finnish clinical microbiology expert and the specimens are manufactured according to the quality standards ISO 9001 and ISO 17043. Before they are sent to the participating laboratories they are tested in three Finnish clinical microbiology laboratories. The scheme of quantitative urine culture consists annually of four rounds, each with two specimens yielding up to 24 specimens in the study period. The specimens were lyophilized microbial suspensions. In the participating laboratory, they were suspended in a 100ml buffer solution to represent a urine specimen and then cultured according to the routine method used in the laboratory.
Based on the microbiology expertise (agreed in advance), the laboratory was allowed to report either only the quantitative culture results of the specimens and a basic evaluation of the significance of growth or it was allowed to report also the microbial identification results and carry out the susceptibility testing. The expected findings in the EQA specimens in the period 2009-2011 are presented in Table 1.
| Expected finding |
Number of microbes (CFU/ml) |
Number of specimens |
| Escherichia coli |
≥105 |
8 |
| Klebsiella sp |
≥105 |
3 |
| Salmonella Virchow |
≥105 |
1 |
| Pseudomonas aeruginosa |
≥105 |
1 |
| Acinetobacter baumannii |
≥105 |
1 |
| Enterococcus sp |
≥105 |
4 |
| Streptococcus agalactiae |
≥105 |
1 |
| Staphylococcus saprophyticus |
≥105 |
1 |
| Aerococcus urinae |
104-5 |
1 |
| Mixed flora (Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa) |
≥105 |
1 |
| Mixed flora (Escherichia coli, Proteus mirabilis, Enterococcus faecalis) |
≥105 |
1 |
| negative |
No growth |
1 |
Table 1: The expected findings in the External Quality Assurance (EQA) urine specimens of Labquality Ltd in the study period 2009-2011.
Culture methods
Based on clinical microbiology textbooks and the guidelines given by a Finnish advisory group [2], urine specimen can be cultured either on dipslide or on agar plate with a 1 μl loop. In this study, 79 of the laboratories used the semiquantitative dipslide method and 246 the plate culture. The plates used were non-chromogenic (CLED or Brolacin, n=169) as well as some commercial chromogenic media designed for urine culture (n=77). The results on microbial growth by both methods were expressed quantitatively in terms of colony forming units /ml (CFU/ml), 103, 103-4, 104-5, >105.
Evaluation of EQA results
EQA results are confidential but according to the Finnish Communicable Disease Act [4], the laboratories are obligated to give them to THL. In this study, instead of asking each laboratory to send the paper reports of their EQA results to THL, they were asked to give permission to the clinical microbiology expert at THL to extract their EQA results directly from the database of Labquality Ltd. The results of laboratories that did not give permission for direct access to the database were excluded from this study. Information on the annual number of routine urinary specimens cultured in each laboratory was collected from THL’s register. The quantitative results were evaluated while taking into account the culture method used (dipslide or agar plate). The evaluation also included the microbial identification results of the laboratories that carried out identification.
Statistical methods
The chi-square test was used to compare the results between dipslide and agar plate users and between the sizes of the laboratories. For the more complex associations, logistic regression analysis was used. The effects as changes in percentages were estimated using the delta method [5]. P<0.05 indicated statistical significance.
Results
Of the 335 participant laboratories, 197 (59%) sent their results for all 24 urinary EQA specimens to Labquality Ltd. The total number of results available for evaluation was 6932. Five per cent of the laboratories did not send their results.
The quantitative culture results were correct in 83% (5745/6932) of the results (Table 2). The data on the method used was available for 6795 (98%) results. Only 10 laboratories with 137 results did not give this information (Table 2). Dipslide was used in 79 laboratories and EQA results were correct in 70% of the 1416 reports. This result differed significantly (p<0.001) from the corresponding results of both the laboratories using non-chromogenic (169 laboratories; 86% of 3618 reports were correct) or chromogenic (77 laboratories; 87% of 1761 reports were correct) media.
| Method |
Dipslide (n=79) |
Plate culture on non-chromogenic medium (n=169) |
Plate culture on chromogenic medium (n=77) |
No information of medium (n=10) |
Total number of results received) (n=335) |
No results Received |
| Total number of results |
1416 |
3618 |
1761 |
137 |
6932 |
352 (5%) |
| Number of correct results |
995 (70%) |
3108*** (86%) |
1529*** (87%) |
113 (81%) |
5745 (83%) |
|
*** p<0.001 compared with dipslide
Table 2: The number of laboratories, results and methods used in EQA schemes of quantitative urine culture (Labquality Ltd) during the period 2009-2011.
Gram-negative rods were present in 14 EQA specimens and the quantitative result was correct in 91% of 3964 results: in 87% of the 827 results for dipslide users and in 93% of the 3137 results for plate users (p<0.001) (Table 4). Escherichia coli were present in eight specimens and the number of bacteria was correctly reported in 93% of the 2250 results. Correct results for other gram-negative bacteria present in six specimens were as follows: Klebsiella sp. in 80% of the 861 results, other gram-negative rods (Salmonella sp, Pseudomonas aeruginosa and Acinetobacter baumannii) in 99% of the 853 results.
Gram-positive bacteria were present in seven EQA specimens and the correct quantitative result was found in 68% of 1994 results: in 37% of the 415 for dipslide users and in 77% of the 1579 results for plate users (p<0.001). Enterococcus sp. was present in four specimens and the number of bacteria was correctly reported in 84% of the 1130 results. Staphylococcus saprophyticus was correctly reported in 89% of the 294 results, Streptococcus agalactiae in 23% of the 296 results and Aerococcus urinae in 31% of the 274 results. The number of bacteria in two EQA specimens with mixed flora (containing several bacterial species) was correctly reported in 66% of the 560 results. The number of correct results was similar in dipslide and plate culture users only in the bacterial groups ‘other gram-negative rods’ and of ‘mixed flora’ (Table 3).
| Microbe (number of specimens) |
All laboratories (correct /total number of results) |
Dipslide (correct/total number of results) |
Plate culture (correct/total number of results) |
p |
| E. coli (8) |
93% (2094/2250) |
87% (400/458) |
96% (1694/1792) |
<0.001 |
| Klebsiella sp (3) |
80% (n=688/861) |
73% (137/189) |
82% (551/672) |
0.004 |
| Other gram-negative rods (P. aeruginosa, Salmonella sp, A. baumannii) (3) |
99% (842/853) |
99% (179/180) |
98 (663/673) |
=0.326 |
| Enterococcus sp (4) |
84% (947/1130)) |
45% (109/241) |
94% (838/889) |
<0.001 |
| S. agalactiae (1) |
23% (67/296) |
4% (3/68) |
28% (64/228) |
<0.001 |
| S. saprophyticus (1) |
89% (262/294) |
58% (34/59) |
97% (228/235) |
<0.001 |
| A. urinae (1) |
31% (86/274) |
13% (6/47) |
35% (80/227) |
=0.003 |
| Mixed flora (2) |
66% (369/560) |
60% (70/117) |
67% (299/443) |
=0.120 |
| Total (gram-negative bacteria) |
91% (3624/3964) |
87% (716/827) |
93% (2908/3137) |
<0.001 |
| Total (gram-positive bacteria) |
68% (1362/1994) |
37% (152/415) |
77% (1210/1579) |
<0.001 |
| Total (all microbes) |
82% (5355/6518) |
69% (938//1359) |
86% (4417/5159) |
<0.001 |
Table 3: The microbes and the percentages of correct results for quantitative culture obtained on dipslide and plate culture. (Results of a specimen without growth and specimens with no information of medium were excluded.)
The type of culture method seemed to have a larger effect (-50% with 95% CI -55.1% - -44.8%) than the bacteria group (grampositive or gram-negative, +6.1% with 95% CI +3.6% - +8.6%). However, there was a significant interaction. This finding led us to analyse more carefully the results and growth of S. agalactiae (present in >105 CFU/ml in the original specimen), given the low number of correct results also in laboratories using plate culture (Figure 1). No growth was reported by 37% of the dipslide users and 4% of the plate users (25/68 vs. 8/228; p<0.001). In addition, 23% of non-chromogenic and 41% of chromogenic media (36/159 vs. 28/69; p=0.006) produced the expected growth of >105.
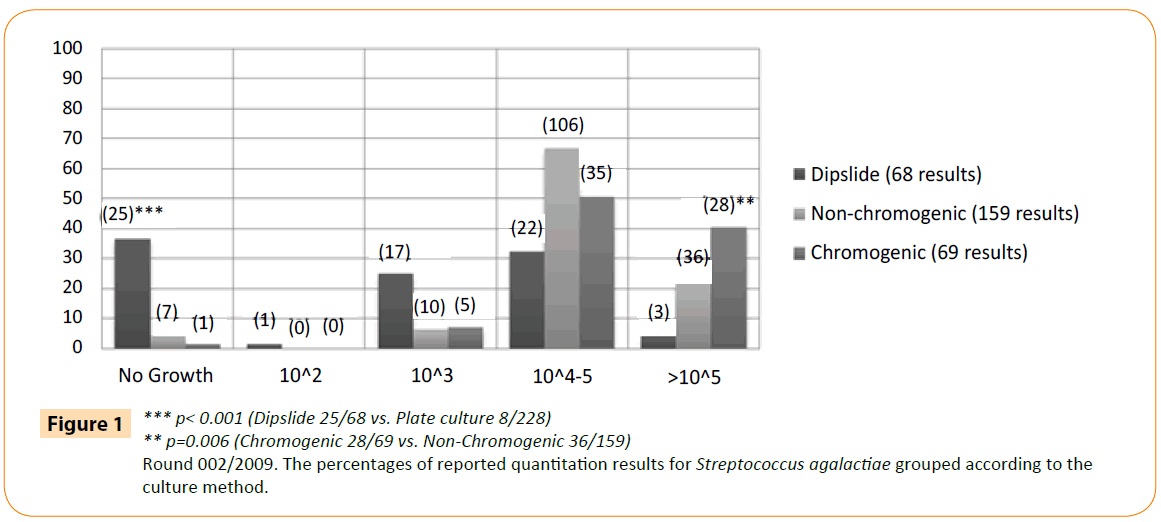
Figure 1: *** p< 0.001 (Dipslide 25/68 vs. Plate culture 8/228)
** p=0.006 (Chromogenic 28/69 vs. Non-Chromogenic 36/159)
Round 002/2009. The percentages of reported quantitation results for Streptococcus agalactiae grouped according to the culture method.
Regardless of whether the size of the laboratory or the culture method used had more influence on the results, the laboratories using dipslide and plate culture were both divided into two groups: laboratories with annual number of routine urine specimens <1000 and those with 1000-10 000 (Table 4). The laboratories with >10 000 specimens were not included, since only one laboratory in this category used dipslide. The EQA results for only gram-positive bacteria were compared, as their correct or incorrect rests seemed to be influenced by the culture method (Table 3). The comparison showed that correct quantitative results for gram-positive bacteria in laboratories using dipslide were less common (97/286; 34%) in those laboratories with <1000 routine urine specimens annually than those with 1000–10 000 annual specimens (54/123; 44%), though the difference was not statistically significant (Table 4). In small laboratories using plate culture, the number of correct results concerning all grampositive bacteria differed significantly (p<0.001, for S. agalactiae p=0.004) from the corresponding results of the dipslide users.
| Media |
Dipslide (correct/total number of results) |
Plate culture (correct/total number of results) |
| Laboratory size Microbe (number of specimens) |
<1000 |
1000–10 000 |
<1000 |
1000–10 000 |
| Enterococcus sp (4) |
43% (71/166) |
53% (37/70) |
92% (252/273)*** |
95% (491/518) |
| S. saprophyticus (1) |
53% (21/40) |
68% (13/19) |
96% (71/74)*** |
97% (130/134) |
| S. agalactiae (1) |
5% (2/44) |
4% (1/24) |
26% (18/70)** |
30% (39/132) |
| A. urinae (1) |
8% (3/36) |
30% (3/10) |
41% (29/71)*** |
32% (42/130) |
| total |
34% (97/286) |
44% (54/123) |
76% (370/488)*** |
77% (702/914)*** |
Table 4: The percentages of correct results of gram-positive bacteria grouped according to the laboratory size (number of annual urine specimens) and culture method.
Identification of uropathogens was performed in 72 of the participating 335 laboratories. Of these, 23 had an annual specimen number >10 000 and they used plate culture in their urine diagnostics. Only two laboratories used dipslide culture. The identification of E. coli was carried out annually in 54 to 65 laboratories, producing 480 identification results (data not shown). The identification was correct in 98% of the results. Other gram-negative uropathogens were identified in 31 to 45 laboratories annually, producing 243 identification results, while for gram-positive bacteria, 17 to 54 laboratories produced 279 identification results annually (data not shown). The success of their identification results varied. There were 10 false results among the gram-negative bacteria and 35 among gram-positive bacteria. The most common cause for a false result was interpreting the growth of gram-positive bacteria as nonsignificant or as mixed flora or reporting negative growth for specimens.
Discussion
The results of the quantitative culture of the 6932 EQA urine specimens in 335 laboratories were evaluated in this study. The specimens were cultured in licensed laboratories using standardised methods. Of these laboratories, only 59% reported the results of all 24 specimens and annually about 5% of the laboratories neglected to report at least one of the rounds. This kind of neglect suggests that the importance of the EQA is not understood in these laboratories. They pay for the specimens but leaving them unreported, thus wasting money and missing the educational opportunity afforded by EQA specimens.
The CFUs of the bacteria were correct in 82% of the reports. There was variation in results based on the bacterial species and the culture method used. Generally, the number of most common uropathogens, that is gram-negative bacteria, was determined more reliable (91% correct) than that of gram-positive bacteria (68% correct). The number of the most common uropathogen, E. coli, was correct in 93%, and Enterococcus sp. in 84% of results, whereas S. agalactiae was correct only in 23% of the results.
The number of bacteria in all EQA specimens, with the exception of one, was high, at least 105CFU/ml. In all incorrect quantitative results, the number of bacteria was lower than 105CFU/ml and, therefore, too low a number of bacteria were reported. For EQA specimens of less than 105CFU/ml, it was not possible in this study to determine how accurate the quantification was. The culture method had a statistically significant influence on the results. Incorrect results (namely too low CFU/ml), not depending on the genus of the bacterium present in the EQA specimen, were reported significantly more often on dipslide than in plate culture (correct results 69% vs. 86%). Similar results have been shown by Aspevall et al. and Morandi et al. [6,7]. However, the opposite results have been obtained in some other studies. Pettersson et al. [8] found higher numbers of bacteria when dipslide was used, leading to over diagnosis of urinary tract infection. Dipslide has also been shown to allow for a reasonable estimate of growth, even if the colony count is as low as 103-4 [9]. In a new study by Bongrad et al., a semiquantitative method was shown to produce false positive results [10]. Laboratories have been shown to succeed better with EQA specimens known to be EQA specimens, than without this knowledge [11]. In routine urine culture, the consequences for the patient are dependent on the interpretation of the bacteria count, while false too-low counts of bacteria may not be reported to physicians.
In the current study, the difference between the methods was most evident with gram-positive bacteria. In the laboratories using dipslide the correct number of gram-positive bacteria was reported in 37% of the results, compared to 77% in plate culture results. Among dipslide users for all other gram-positive bacteria except S. agalactiae, the percentage of correct results was higher if the annual number of routine specimens was also higher (at least 1000/year). However, the higher number of routine specimens did not improve the results statistically significantly. Also, among the plate culture users, the percentage of correct results was not dependent on the annual number of routine urine cultures in the laboratory.
Given the simpler method of using the dipslide, this culture is preferred in small laboratories with less than 1000 annual specimens. According to our results, the plate culture gave significantly more reliable quantitative results also in these laboratories. Differences in quantitative results were not due to the medium, since the CLED medium is used both in plate culture and dipslide. The medium is designed to support the growth of the most common uropathogens, especially gram-negative rods. This may be the reason for the poor growth of gram-positive bacteria compared to gram-negative bacteria reported by all laboratories, leading to a lower percentage of correct results (68% vs 91%). In addition to incorrect CFUs/ml for gram-positive bacteria, there were also a high number of laboratories who reported the negative results for specimens. Thus, probably due to the small colony size and weak growth of gram-positive bacteria, they were undetected and hence not reported.
In this study, only one EQA specimen contained S. agalactiae. Despite high concentration of S. agalactiae in the specimen, 37% of the laboratories using dipslide reported the specimen result as negative, compared to 6% of those using plate cultures. The poor growth of S. agalactiae on dipslide has been observed also in other studies [6,7,12]. If the phenomenon is true, it means that in real life, a urinary tract infection or colonisation caused by gram-positive bacteria is generally not detected. In particular, it is most important to be aware of whether dipslide is recommended in the detection of S.agalactiae in the urine specimens of pregnant women [13]. In clinical microbiology textbooks and also in European Urinalysis guidelines [14], it is recommended to use a non-selective medium, such as blood agar, in addition to the selective medium, such as CLED, in urine culture. However, this recommendation has not been routinely implemented in urine culture in Finland. According to this study urine culture including blood agar should be available in special cases like; routine culture remains negative and the patient has the symptoms of urinary tract infection or patient is pregnant.
Plate culture, mostly on non-chromogenic plate, was the most common method used in 50% of the 335 laboratories in this study. To improve the detection and identification of uropathogens, chromogenic media have been introduced. Chromogenic media were first reported as being taken into use in 2005 in two laboratories participating in EQA schemes for urine culture. The use of chromogenic media has gradually increased in laboratories and during this study period (2009-2011), there were already 77 laboratories using chromogenic media. Chromogenic media were shown to be equal to non-chromogenic media in the determination of the number of bacteria. However, interestingly, chromogenic media supported the abundant growth of S. agalactiae significantly better than non-chromogenic media.
Urine culture is the most common microbiology culture performed in the laboratories from the clinical specimens collected from ill subjects. In this study, most of the laboratories (90%) were not clinical microbiology laboratories, but small laboratories either inside health centres or in the private sector. The annual number of routine urine specimens handled in these laboratories was less than 10 000, while in larger laboratories it was over 10 000. Due to the high number of annual urine culture specimens, laboratories are constantly seeking ways to improve methods to process urine specimens. Culture automates have been introduced to dispense the specimen onto culture plates [15,16]. In addition to reducing labour demands in the laboratory, they may diminish some of the inaccuracies found in the manual loop culture [17]. Moreover, in order to reduce the workload of culturing, the use of optical instruments, that is flow cytometers, has been used to screen for negative urine specimens. This has been shown to reduce the need to culture urine samples by almost 65% [18].
These innovations can only be taken into use in bigger laboratory units, with a high number of specimens. The basic work-urine culture and defining the number and considering the significance of bacteria may often still need to be done in small laboratories. New microbiological methods are currently under development, but they are not yet ready for routine with the high number of specimens [19].
Our results showed that in spite of the numerous training sessions already given on urine culture in Finland, more knowledge is still needed to also detect and recognise the less common uropathogens, such as A. urinae and S. agalactiae. These results also emphasise that a clinical microbiological investigation is not only the performance of a laboratory test but an analysis of a specimen leading to a diagnosis for a patient. Since the number of microbes detected and reported is influenced by the culture method and the criteria used in the laboratory, physicians should have close contact with the diagnostic laboratory in order to make a correct diagnosis of urinary tract infection for a patient. Diagnostic laboratories should also be well informed when special culture like use of blood agar is relevant. This also means that administrators should make decisions that enable an environment where contacts between physicians and the laboratory are possible.
8449
References
- Stamm WE (1983) Measurement of pyuria and its relation to bacteriuria. Am J Med 75: 53-58.
- Koivula T, Grönroos P, Gävert J, Icen A, Irjala K, et al. (1990) Basic urinalysis and urine culture: Finnish recommendations from the working group on clean midstream specimens. Scand J Clin Lab Invest Suppl 200:26-33
- OjanenTarja (2002) Is Labquality’s recommendation for urine culture implemented the results of the survey.
- https://www.doria.fi/bitstream/handle/10024/72549/AnnalesD995Nikula.pdf
- Cox DR, Snell EJ (1989) Analysis of Binary Data, Second Edition. Chapman and Hall/CRC.
- Aspevall O, Kjerstadius T, Lindberg L, Hallander H (2000) Performance of Uricult Trio assessed by a comparison method and external control panels in primary healthcare. Scand J Clin Lab Invest 60: 381-386.
- Morandi PA, Mauris A, Deom A, Rohner P (2007) External quality control results of urine dip-slide devices. DiagnMicrobiol Infect Dis 57: 235-241.
- Pettersson L, Kinnunen K, Forsum U (1995) Risk of overdiagnosis of urine cultures. Lakartidningen 92: 2195-2196.
- Frimodt-Møller N, Espersen F (2000) Evaluation of calibrated 1 and 10 ul loops and dipslide as compared to pipettes for detection of low countbacteriuria in vitro. APMIS 108: 525-530
- Bongard E, Frimodt-Møller N, Gal M, Wootton M, Howe R, et al. (2015) Analytic laboratory performance of a point of care urine culture kit for diagnosis and antibiotic susceptibility testing. Eur J ClinMicrobiol Infect Dis 34: 2111-2119.
- Kumasaka K, Kawano K, Yamaguchi K, Igari J, Minowa K, et al. (2001) A study of quality assessment in clinical microbiology performance of independent laboratories in Tokyo: 18-year participation in the Tokyo Metropolitan Government External Quality Assessment Program. J Infect Chemother 7: 102-109
- Jokipii AM, Jokipii L (1979) Recognition of group B streptococci in dip-slide cultures of urine. J ClinMicrobiol 10: 218-221.
- Mignini L, Carroli G, Abalos E, Widmer M, Amigot S,et al. (2009) Accuracy of Diagnostic Tests to Detect Asymptomatic Bacteriuria in Pregnancy. Obstetrics and Gynecology 113: 346-352.
- Høiby N, Bjarnsholt T, Moser C, Bassi GL, Coenye T, et al. (2014)ESCMID guideline for the diagnosis and treatment of biofilm infections 2014.Scand J Lab invest 60:1-96
- Sharon S, Bourbeau PP (2015) Impact of introduction of the BD KiestraInoqulA on urine culture results in a hospital clinical microbiology laboratory. J ClinMicrobiol53:1736-1740
- Hirvonen J,Liimatainen O, Vuento R(2014) Comparison of Copan WASP plating system and manual culture in streaking of urine specimens. ECCMID
- Albers AC, Fletcher RD (1983) Accuracy of calibrated-loop transfer. J ClinMicrobiol 18: 40-42.
- Jolkkonen S, Paattiniemi EL, Kärpänoja P, Sarkkinen H (2010) Screening of urine samples by flow cytometry reduces the need for culture. J ClinMicrobiol 48: 3117-3121.
- Wu F, Dekker C (2016) Nanofabricated structures and microfluidic devices for bacteria: from techniques to biology. ChemSoc Rev 45: 268-280.






