Keywords
COVID 19; Infection; Babies; Understanding; Recombination
Introduction
In late December 2019, in Wuhan City (Hubei Province, China), a novel coronavirus (CoV) named ‘ ‘ 2019-nCoV ’ ’ responsible for severe acute respiratory syndrome was declared by the World Health Organization (WHO) [1]. Because of the international travel, the virus spread from this city to the rest of the world, the 11 May, 2020 we recorded 4006257 confirmed cases and 278892 confirmed deaths in 2020 [2].
Due to the great debate of CoV19 origin and development, we think that we have a limited knowledge. Coronaviruses are able to survive in many environments through an acquired mutation and recombination with relative ease to alter host range and tissue tropism efficiently [3]. The evolution and diversification of viruses is due to virus cross-species transmission among hosts, appearing in many instances to be preferential to coevolving within an initial host [4].
Literature Review
The coronavirus pandemic has stirred up a great debate about the selective age for this virus. Many researchers indicated that virus infection is not selective in age, as it was reported even in a 1-month-old infant [5]. Among the 44 672 confirmed cases, 77.8% are between 30 and 69 years old and 51.4% are male [6]. Furthermore, published data suggest that babies and children are less likely to be infected than adults and older one.
Unlike infected adults, children consistently make up <2% of total case numbers in reported ones as discussed here. Thus, the determination of the spectrum of disease in children is subject to many limitations especially for developing countries. Consistent with RCPCH Research and evidence team reports [7], it has been shown that children under the age of 18 and infected with COVID-19 represent only 2% of total cases worldwide. In another epidemiological study [8], it has been reported that few patients of children and infants were infected compared to patients aged between 35 and 55 years. Based on other data published so far [9], it has been reported that COVID-19 was associated with age. Many reports found that children younger than nine years accounted for just 1% whereas children aged between ten and 19 accounted for 5.2% of cases.
Data from CDC (Centers for Disease Control and Prevention) suggest that most COVID-19 cases in children are not severe, serious COVID-19 illness resulting in hospitalization still occurs in this age group [10]. Furthermore, Lu et al. [11] reported that pediatric COVID-19 cases were less severe than cases in adults.
Disease characteristics among pediatric patients in the United States have not been described [12] despite the three cases of CoV19 positive children who died in New Jersey [10]. In morbidity and mortality weekly report [9], American children infected by COVID-19 under 18 years old represented only 1.7% of the whole population. The CDC report have shown that infants <1 remain underrepresented among COVID-19 cases in patients of all ages (393 of 149,082: 0.27%) [10].
According to CDC data, children aged <1 and 1-4 years presented 15%, 11%, respectively [10]. Notably, Lavezzo et al. [13] outcome revealed no cases among the 374 children under ten years of the city of Vo' (Province of Padua, Italy). Accordingly, this outcome might underestimate the reality because of the reported pediatric cases and their links with the CoV19 infection since most cases reported among children and babies to date have not been severe. So, reasons for any potential difference in COVID-19 incidence or severity between ages remain unclear. The RCPCH research indicated a low rate of 0.01% (equivalent to one in 10,000 cases) of death in children [7]. Other data from China and Italy indicated that children represented approximately 5% of cases and less than 1% of admissions to hospital [14]. Few cases were reported under 5 or 6 years. Currently, it is known that 5.6% was the result of the largest review of children with severe COVID-19 (112/2143 Chinese cases) [15].
Are babies less vulnerable and largely spared from the effects of the SARS-CoV-2 or related coronavirus?
Understanding how and why babies are not susceptible to infection with this virus is critical. The clinical outcomes for babies and CoVID-19 are limited, no estimation was given. Here, we are highlighting how babies are less vulnerable to COVID-19 ’ s effects or attacks. We explain in part recent findings on the CoV19 recombination and immunization.
According to Bi et al. [16] deaths in children due to COVID-19 have been extremely rare (0.01%), for babies it is not yet clear and many questions are unanswered. Basically, two factors are presented here: ACE-2 receptors and babies’ vaccination, providing protection against CoV19 infections.
Importantly, the virus needs a receptor on the target cell to be known and to start his life cycle. Initial encounters between a virus and a host cell are mediated through viral surface components, either membrane glycoproteins or sites on a viral capsid [17]. The ligands that are on the target cell surface can be glycolipid and/or of glycoprotein nature [18].
A several cell surface components were described and identified, certain viruses use single molecular species as receptors such as CD155 for poliovirus [19,20], the low-density lipoprotein receptor (LDLR) for human rhinovirus 2 [21], and dendritic cell specific intercellular adhesion molecule-3- grabbing nonintegrin (DC-SIGN) for the phleboviruses (a subgroup of bunyaviruses) [22]. While, more than one molecular species are used as receptors: angiotensinconverting enzyme (ACE) or liver-SIGN (L-SIGN) for SARS coronavirus [23,24]; and scavenger receptor-B2 (SR-B2) or Pselectin glycoprotein ligand-1 (PSGL-1) for enterovirus 71 [25]. For other viruses like Human immunodeficiency viruses (HIVs), the process is more complex. Multiple cell surface components are required and receptors are exhibited dependency that involves engagement with at least two distinct plasma membrane components [19].
Angiotensin-converting enzyme 2 (ACE2) was found to be an efficient receptor for the S glycoprotein of SARS-CoV [26,27]. As reported in several researches, children have less ACE-2 receptors in their lower airways (lungs) than in their upper airways, that is why they have thankfully been largely spared from the COVID-19 risks and effects.
As reported by Peiris et al. [28] the SARS-CoV virus causes atypical pneumonia with diffuse alveolar damage with an overall mortality of ≈10% that ranges from 0% in children and 50% in persons over 65. Thus, the number of reported COVID-19 cases in babies remains low and no deaths cases were reported. They are less vulnerable to COVID-19 effects, but they can still be infected. The public health researchers indicated that children are not the face of this pandemic. The first question to be asked is why the virus seems to affect children and babies differently?
We should ask ourselves whether the low numbers of child infections recorded is due to the low numbers of children being exposed or being infected. Several questions need to be addressed within the stronger transmission and propagation of the new CoVID19 virus. First, why is this virus more transmissible than other CoV that cause severe respiratory disease (SARS-CoV and MERS-CoV)? Is it a developed virus or a mutated one? Second, SARS-CoV and MERS-CoV infection in babies is less commonly reported and sometimes absent, is the virus mutating to ovoid babies? Is because of their immunization (vaccination schedules)?
It is noteworthy that via the vaccination, babies are protected from many immunogens. It was reported that a child who receives all the recommended vaccines in his immunization schedule may be exposed to up to 320 antigens through vaccination by the age of 2 [29]. Antibodies seem to provide a protective immunity for babies, as discussed in detail in the following sections.
CoV19 and vaccination schedule
We refer to the vaccination schedule to give our opinion and to explain the link between the CoV19 and the other ARN viruses. Genetic recombination between strains of vaccine / vaccine or vaccine / wild poliovirus, and between enterovirus is a phenomenon known for several years [30], the biological consequences of these genetic exchanges are responsible for several epidemics. Furthermore, it was suggested that genetic modifications contribute significantly in the evolution of coronaviruses and the acquisition of a new or more virulent phenotype responsible for the CoV19 pandemic.
Moreover, CoV19 seems to avoid babies because they are immunized against paramyxovirus, morbillivirus, rhinovirus, enterovirus and influenza virus during the first 5 years. They have high level of antibodies against viruses in their immune systems. The concept of immunosenescence is not put into play in this review since young adult are also victim of CoV19.
IgM was the first produced antibody in the case of a primary immune response, without isotype switching and with the constant kinetic: IgM-IgG; IgM-IgA; IgM-IgE. A significant amount of antibodies in a baby’s immune system will allow the SELF to face several antigens. The outcome of this active (acquired) immunization depends on the nature of the vaccine (live-attenuated, inactivated or a subunit) or on these immunogenic properties.
In addition to BCG vaccine, ten antigenic fragments: Diphtheria (Diphtheria toxoid), Tetanus (inactivated tetanus toxin), Poliomyelitis (Inactivated polio vaccine), Pertussis (DTaP: diphtheria, tetanus, and acellular pertussis), Meningitis due to Haemophilus: influenzae b (polysaccharide vaccine), pneumonia and pneumococcal septicemia (polysaccharide vaccine), Hepatitis B (Subunit/conjugate), Measles, mumps, rubella (MMR combined vaccine) (Live, attenuated vaccine) are administered during the first months (2, 4 and 11) and the 12 months.
The best evidence supporting our opinion comes from Netea et al. [31] study, the BCG vaccine and some other live vaccines are able to induce metabolic and epigenetic changes that enhance the innate and the specific immunity already controlled by LTCD4. Furthermore, Curtis et al. [32] reported that viraemia after SARS-COV-2 exposure is reduced in the presence of BCG vaccine with consequent less severe COVID-19. On 11 April 2020, WHO updated its ongoing evidence review of the major scientific databases and clinical trial repositories for COVID-19 and BCG vaccine. This update was done according to three preprints in which the authors have tried to explain the link between the incidence of COVID-19 cases in countries and the BCG vaccine. We felt that several aspects of this link need further researches since the WHO recommend only neonatal BCG vaccination.
Additionally, during natural breast feeding babies are passively immunized (passive immunity). Maternal antibodies (isotype IgA) are transferred from breast milk or colostrum’s to the GALT system (Gut Associated Lymphoid Tissue). The acquired immunity presents an anti-infectious role and enhances immune functions such as opsonization, neutralization and activation of the complement. An earlier study by Van de Perre [33] showed the presence of antiidiotypic antibodies in the maternal milk. These molecules are capable of enhancing infant antibody response especially for mucosal portal of entry. In the same study, antirotaviral IgA can be detected in stools of breast-fed but not bottle-fed neonates.
The above studies provide insight into the importance of babies’ vaccination. Antibodies are playing a fundamental role in the humoral immunity (LTB) and indirectly are orienting the cellular response (LTH (TCD4) and CTLs (TCD8)). So, with this antibodies ’ diversity, each isotype or molecule will target specifically in either binding sites on outer proteins or nucleic acid. Therefore, from all what was mentioned above, it seems clear that immunizing babies presents a protective barrier against CoV19. The mentioned evidence has supported the role of the active immunization (vaccination) to protect babies from CoV19 infection.
Moreover, the immune memory is generated by vaccination; antibodies are secreted continuously to induce rapid recall responses. Our discussion corroborates with Cao et al. [34] study about SARS-CoV-2 infection in children. According to Zhang [35], leukocytes and lymphocytes levels were normal and no depletion occurred within CoV19 virus.
The β-coronaviruses lineages and CoV19
As mentionned by Zhou et al. [36] Coronaviruses (CoVs) possess the largest known RNA genomes identified so far typically ranging from 27 to 32 kb [37]. Coronaviruses (CoVs) are from Nidovirales order and belong to Coronaviridae family and Coronavirinae subfamily [38], which is divided into four genera, α-coronavirus, β-coronavirus, δ-coronavirus and γ- coronaviruses. The most diversified genus is the β- coronaviruses with 4 lineages (A, B, C and D). Previously, it was demonstrated that Alpha-CoVs and beta-CoVs circulate in mammalian hosts, whereas avian species are infected by gamma coronaviruses. Delta coronaviruses infect both avian and mammalian species [39].
In Wu et al. [40] commentary, six kinds of human CoVs are highlighted. Representative alpha coronaviruses include human coronavirus NL63 (HCoV-NL63), porcine transmissible gastroenteritis coronavirus (TGEV), enteropathogenic porcine epidemic diarrhea virus (PEDV), and porcine respiratory coronavirus (PRCV). The known severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV), mouse hepatitis coronavirus (MHV), bovine coronavirus (BCoV), and human coronavirus OC43 belong to the Beta coronavirus genus [31]. Avian infectious bronchitis coronavirus (IBV) [40] and porcine delta coronavirus (PdCV) represent gamma coronaviruses and delta coronaviruses, respectively [41].
Previous reports showed seven CoVs infecting humans (HCoVs), represented by HCoV-229E, HCoV-NL63 in the α- coronaviruses and HCoV-OC43-HCoV-HKU1 in the β- coronaviruses lineage A. In addition, SARS-CoV and SARS-CoV-2 belong to β- coronaviruses lineage B and MERS-CoV to β- coronaviruses lineage C (Figure 1).
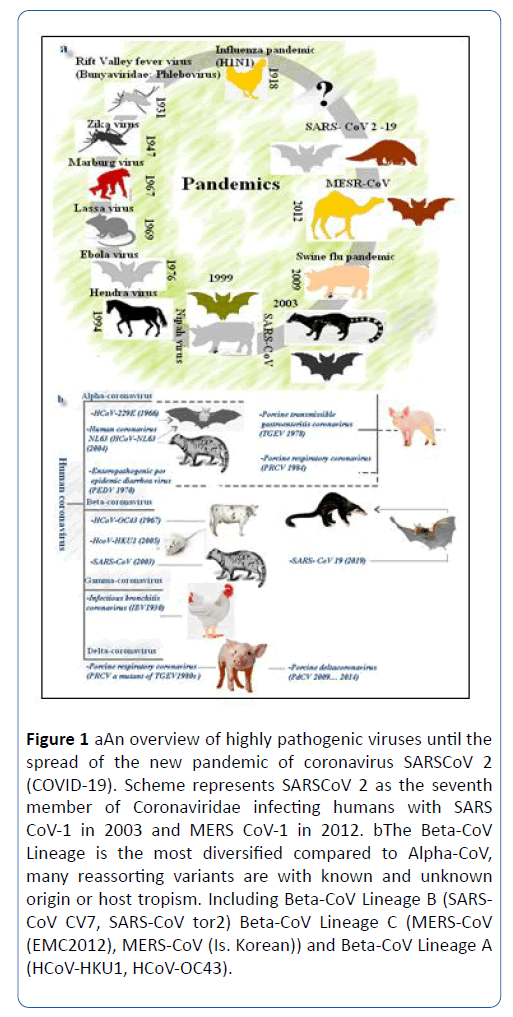
Figure 1: a. An overview of highly pathogenic viruses until the spread of the new pandemic of coronavirus SARSCoV 2 (COVID-19). Scheme represents SARSCoV 2 as the seventh member of Coronaviridae infecting humans with SARS CoV-1 in 2003 and MERS CoV-1 in 2012. b. The Beta-CoV Lineage is the most diversified compared to Alpha-CoV, many reassorting variants are with known and unknown origin or host tropism. Including Beta-CoV Lineage B (SARSCoV CV7, SARS-CoV tor2) Beta-CoV Lineage C (MERS-CoV (EMC2012), MERS-CoV (Is. Korean)) and Beta-CoV Lineage A (HCoV-HKU1, HCoV-OC43).
Several recent excellent reviews delve into the knowns and the unknowns of HCoV origin in terms of reservoir species, amplification host, and, more generally, of CoV ecology [40]. Interestingly, Vijgen et al. [42] reported in an absolute quantitation of human Coronaviruses study that HCoVs are recognised as one of the most rapidly evolving viruses owing to its high genomic nucleotide substitution rates and recombination. Other studies found a significant association of urbanization and poultry farming in the expedition of HCoVs evolution. Like many other viruses, it seems from the available data of Jones et al. [43] that this emergence is linked to agricultural intensification and environmental change. All these factors and change have permitted a frequent mixing of species and facilitated the crossing of species barrier and genomic recombination [43].
Plasticity of the coronavirus (CoVs) genome
Thus, the significant plasticity of the coronavirus (CoVs) genome makes these viruses agents with high evolutionary potential. CoVs are prone to genetic evolution through accumulation of point mutations and homologous recombination among members of the same genus [44]. There are numerous reports of CoV genomes; in which the evidence favors the view that they display high plasticity in terms of gene content and recombination. Forni et al. [45] research on molecular evolution of human coronavirus has provided novel insight into it genome. This genetic instruction expands the sequence space available for adaptive mutation, and the spike protein can adapt new strategy with relative ease to interact and bind to many receptors on target cells.
Genome expansion in CoVs is believed to be at least partially mediated by increased replication fidelity. Given its continuous exposure to the host, the mutation rate for CoVs differs. Possibly depending on the phase of CoV adaptation to target hosts. Early study demonstrated that these viruses may possess an unusually high replication fidelity [46] compared to other RNA viruses. Recent research published the genome size of coronaviruses, which ranges between approximately 26,000-32,000 bases and includes a variable number (from 6 to 11) of open reading frames (ORFs) [47]. The first one (ORF1a and ORF1b) encodes the replicas proteins with a great extension (2/3 of the genome) and the remaining ORF1 (1/3) encodes structural proteins: S (spike protein), E (envelope protein), M (membrane protein) and N (nucleocapsid protein) [48].
It is interesting to note that the high mutation rates in RNA viruses may help researchers to track the spread and the evolution of SARS-CoV-2 [49] especially when the host is a child or baby. It was reported that Coronaviruses likely have lower mutation rates than other RNA viruses because of an inherent capacity for some proof-reading activity due to a 30- to-50 exoribonuclease [50]. Nowadays, it seems that this conclusion has changed since this virus had acquired high mutation rates and had preferentially affected adults at the moment and not babies. The evidence to date suggests that very few children develop severe Covid-19 symptoms or can be asymptomatic.
Contrastingly, researchers have no answers or explanations regarding the significant number of amino acid substitutions between the 2019-nCoV and SARS or SARS-like CoVs. They discovered six mutations occurred on the other region of the receptor-binding binding motifs that directly interact with human receptor ACE2 protein [39]. This data has contributed extensively to explain the host tropism and transmission property of the 2019-nCoV compared to SARS-CoV but it is essential to gather many studies on children or babies.
The common feature of these viruses is that they are positive and negative ARN viruses. However, coronaviruses are quite distinct since they are positive-sense RNA viruses, which undergo recombination at high frequency. Chen and Li [51] had proposed that CoVs (and other viruses) stole additional genes from their hosts. CoV genomes do not only evolve by gene gains and losses, but also via subtler changes that modify protein sequences, and recombination has an important role in reassorting variants [45].
As mentioned above, many molecular pathways are considered to be responsible for the CoV evolution although the evolutionary history of CoV19 is controversial. Recent studies indicated that genomes not only evolve by gene gains and losses, but also via subtler changes that modify protein sequences [45]. Other researchers assumed that recombination events have an important role in reassorting variants and the genetically engineered modifications may affect the CoV evolution without any advantageous for health research investigations.
Furthermore, Forni et al. [45] reported that artificial selection can lead to unintended changes in viral genomes. Their most important conclusion is that such changes most likely result from passages in culture that, on one hand, relieve the virus from pressures exerted in vivo (e.g. by the host immune system) and, on the other hand, derive from viral adaptation to the in vitro system.
With the genesis of CoV19 variants vaccines will continue to play an important role in maintaining babies' health. The above discussion suggests that vaccination as an acquired immunization might have a protecting role for babies against coronavirus, these researches warrant need further discussions.
Discussion and Conclusion
The recent reports on the identification of the cause of the current CoV19 pandemic does not provide much clarity but rather adds a confusion regarding the nature of the CoV19 and the exposed population. Many published data had swung the debate to favor one hypothesis over another about CoV19 origin.
There is still much to understand how cross-species transmission of CoVs and recombination occurs, especially in children and babies. The scholarly community should conduct further researches to identify the possible recombination of CoV19 with other ARN viruses. These researches may also be of appreciable importance to explain the fact that the proportion of children developing Covid-19 symptoms was much lower compared to adults and the virus has affected adults at the moment than babies with other reasserting variants and other ways. Herein, we highlighted how vaccination and immune memory in babies come into play.
Declaration of Interest
The author declares no conflicts of interest.
Acknowledgement
Not applicable.
28347
References
- Wang D, Hu B, Hu C, Zhu F, Liu X, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. JAMA. 323: 1061-1069.
- https: //www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/
- Li WH, Wong SK, Li F, Kuhn JH, Huang IC, et al. (2006) Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2–S-protein interactions. J Virol. 80: 4211-4219.
- Geoghegan JL, Duchêne S, Holmes EC (2017) Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 13: e1006215.
- Wj Guan, ZY Ni, Y Hu, Liang WH, Chun-quan OU, et al. (2020) Clinical characteristics of 2019 novel coronavirus infection in China. N Engl J Med. 382: 1708-1720.
- Zhang Y (2020) The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Chinese Journal of Epidemiology. 41: 145-151.
- RCPCH Research and Evidence team (2020) COVID-19-research evidence summaries. Royal College of Paediatrics and Child Health.
- COVID-19 National Emergency Response Center, Epidemiology and Case Management Team, Korea Centers for Disease Control and Prevention (2020) Coronavirus Disease-19: The First 7,755 Cases in the Republic of Korea. Osong Public Health Res Perspect. 11: 85-90.
- CDC COVID-19 Response Team (2020) Coronavirus Disease 2019 in Children - United States, February 12-April 2, 2020. MMWR Morb Mortal Wkly Rep. 69: 422-426.
- Lu X, Zhang L, Du H, Zhang J, Qu J, et al. (2020) SARS-CoV-2 infection in children. N Engl J Med. 382:1663-1665.
- https: //www.weforum.org/agenda/2020/05/syndrome-covid19-children-new-york coronavirus/
- Lavezzo E, Franchin E, Ciavarella C, Dannenburg GC, Barzon L, et al. (2020) Suppression of COVID-19 outbreak in the municipality of Vo, Italy. medRxiv.
- Ludvigsson JF (2020) Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr. 109: 1088-1095.
- Dodi I, Castellone E, Pappalardo M, Rubini M, Veronese P, et al. (2020) SARS-CoV-2 infection in children in Parma. Acta Biomed. 91: 214-215.
- Bi Q, Wu Y, Mei S, Chenfei Ye, Xuan Zou, et al. (2020) Epidemiology and Transmission of COVID-19 in Shenzhen China: Analysis of 391 cases and 1,286 of their close contacts. Lancet Infectious Diseases.
- Marsh MA (2006) Helenius Virus entry: Open sesame. Cell. 124:729-740.
- Vlasak M, Goesler I, Blaas D (2005) Human rhinovirus type 89 variants use heparan sulfate proteoglycan for cell attachment. J Virol 79: 5963-5970.
- Grove J, Marsh M (2011) The cell biology of receptor-mediated virus entry. J Cell Biol. 195: 1071-1082.
- Mendelsohn CL, Wimmer E, Racaniello VR (1989) Cellular receptor for poliovirus: Molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 56: 855-865.
- Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, et al. (1994) Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc Natl Acad Sci USA. 91: 1839-1842.
- Lozach PY, Kühbacher A, Meier R, Mancini R, Bitto D, et al. (2011) DC-SIGN as a receptor for phleboviruses. Cell Host Microbe. 10: 75-88.
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, et al. (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 426: 450-454.
- Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, et al. (2004) CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 101: 15748-53.
- Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, et al. (2009) Scavenger Receptor B2 is a Cellular Receptor for Enterovirus 71. Nat Med. 15: 798-801.
- Wang P, Chen J, Zheng A, Nie Y, Shi X, et al. (2004) Expression cloning of functional receptor used by SARS coronavirus. Biochem Biophys Res Commun. 315: 439-444.
- Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, et al. (2018) Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep. 8: 15701.
- Peiris JS, Yuen KY, Osterhaus AD, Stohr K (2003) The Severe Acute Respiratory Syndrome. N Engl J Med. 349: 2431-41.
- https: //www.cdc.gov/vaccinesafety/concerns/multiple-vaccines-immunity.
- Cuervo N, Guillot S, Romanenkova N, Combiescu, M, Combiescu AA, et al. (2001) Genomic features of intertypic recombinant Sabin strains excreted by primary vaccinees. J Virol. 75: 5740-5751.
- Netea MG, Dominguez-Andres J, Barreiro LB et al. (2020) Defining trained immunity and its role in health and disease. Nat Rev Immunol. 20: 375-388.
- Curtis N, Sparrow A, Ghebreyesus TA, Netea MG (2020) Considering BCG vaccination to reduce the impact of COVID-19. The Lancet. 395: 1545-1546.
- Van de Perre P (2003) Transfer of antibody via mother’s milk. Vaccine. 21: 3374-3376.
- Cao Q, Chen YC, Chen CL, Chiu CH (2020) SARS-CoV-2 infection in children: Transmission dynamics and clinical characteristics. J Formos Med Assoc. 119: 670-673.
- Zhang YP (2020) The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Chin J Epidemiol. 41: 145e51.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. (2020) pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579: 270–273.
- Li F (2016) Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol. 3: 237-261.
- Masters PS, Perlman S (2013) Coronaviridae. In: Fields virology. Knipe DM, Howley PM (eds.), 6th edn, Philadelphia: Lippincott Williams & Wilkins. Pp: 825-58.
- Woo PCY, Lau SKP, Lam CSF, Lau CCY, Sang AKLT, et al. (2012) Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 86: 3995-4008.
- Wu A, Peng Y, Huang B, Ding X, Wang X, et al. (2020) Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 27: 325-328.
- Boley PA, Alhamo MA, Lossie G, Yadav K, Vasquez-Lee M, et al. (2020) Porcine Deltacoronavirus Infection and Transmission in Poultry, United States. Emerg Infect Dis. 26: 255-265.
- Vijgen L, Keyaerts E, Moës E, Maes P, Duson G, et al. (2005) Development of One-Step, Real-Time, Quantitative Reverse Transcriptase PCR Assays for Absolute Quantitation of Human Coronaviruses OC43 and 229E. J Clin Microbiol. 43: 5452-5456.
- Jones BA, Grace D, Rushton J, Said MY, McKeever D, et al. (2013) Zoonosis emergence linked to agricultural intensification and environmental change. Proc Natl Acad Sci USA. 21: 8399-8340.
- Enjuanes L, Brian D, Cavanagh D, Holmes K, Lai MMC, et al. (2000)Coronaviridae. In: Virus taxonomy. Van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, et al. (eds.) Classification and nomenclature of viruses. New York: Academic Press. Pp: 835-839.
- Forni D, Cagliani R, Clerici M, Sironi M (2017) Molecular Evolution of Human Coronavirus Genomes.TrendsMicrobiol 25: 35-48.
- Vega VB, Ruan Y, Liu J, Lee WH, Wei CL, et al. (2004) Mutational dynamics of the SARS coronavirus in cell culture and human populations isolated in 2003. BMC Infect Dis. 4: 32.
- Song Z, Xu Y, Bao L, Zhang L, Yu P, et al. (2019) From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses. 11: 59.
- Kopecky-Bromberg SA, Martı´nez-Sobrido L, Frieman M, Baric RA, Palese P (2007) Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists. J Virol. 81: 548-557.
- Grubaugh ND, Ladner JT, Lemey P, Pybus OG, Rambaut A, et al. (2019) Tracking virus outbreaks in the twenty-first century. Nat Microbiol. 4: 10-19.
- Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, et al. (2006) Discovery of an RNA virus Virus 3'->5'exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA 103: 5108-5113.
- Chen L, Li F (2013) Structural analysis of the evolutionary origins of influenza virus hemagglutinin and other viral lectins. J Virol. 87: 4118-4120.
- Anisimova M, Nielsen R, Yang Z (2003) Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics.164: 1229-1236.







