Research Article - (2022) Volume 16, Issue 2
Antibiotic susceptibility pattern of coagulase positive Staphylococcus aureus isolated from patients admitted at Tikur Anbessa specialized Hospital, Addis Ababa
Temesgen Eticha1*,
Tewodros Tamire2 and
Temesgen Bati Gelgelu3
1Department of medical laboratory sciences, college of health sciences and medicine, Ethiopia
2Tikur Anbessa specialized hospital, Addis Ababa, Ethiopia
3School of public health, college of health sciences and medicine, Wolaita Sodo University, Wolaita S, Ethiopia
*Correspondence:
Temesgen Eticha, Department of medical laboratory sciences, college of health sciences and medicine,
Ethiopia,
Tel: +251-926009489,
Email:
Received: 08-Dec-2021, Manuscript No. iphsj-21-11932;
Editor assigned: 10-Dec-2021, Pre QC No. Preqc No.11932;
Reviewed: 07-Feb-2022, QC No. QC No.11932;
Revised: 12-Feb-2022, Manuscript No. iphsj-21-11932 (R);
Published:
21-Feb-2022, DOI: 10.36648/1791-809X.16.2.911
Abstract
Background: Coagulase-positive S. aureus is among the most ubiquitous and dangerous human Pathogens, for both its virulence and its ability to develop antibiotic resistance. Development of Antimicrobial resistance has limited treatment options against infections due to this pathogen. The aim of this study was to assess the Antimicrobial agent’s susceptibility of coagulase-positive S. aureus isolated from patients admitted at Tikur Anbessa specialized hospital, Addis Ababa Ethiopia
Methods: The study was part of a cross sectional study conducted among 413 admitted patients Suspected of HAI to assess the prevalence of Staphylococcus aureus and associated risk factors. Participants were recruited using consecutive convenient sampling technique. Antimicrobial Agents susceptibility were tested on (n=160) coagulase positive S. aureus and (n=57) MRSA Isolates to panels of 9 antimicrobial agents using disc diffusion assay QC was performed to Check the quality of medium. Each new lot was quality controlled before use for testing the Staphylococcus aurous using standard strains (ATCC25923). Collected data were entered and analysed using SPSS version 24.
Result: From the total Coagulase positive S. aureus’s (n=160), 142(88.8%) isolates were Resistant to Penicillin 143(89.4%) followed by Erythromycin 93(58.1%), Gentamicin 87(54.4%), Ciprofloxacin 89(55.6%), Cefoxitin 57(35.6%), Trimethoprim Sulfamethoxazole 69(43.1%), Clindamycin 9(5.6%), ceftriaxone and Augmentin each 37(35.6%). All 57 MRSA strains were 100% resistant to Penicillin, Erythromycin, Augmentin, Cefoxitin and Ceftriaxone. Whereas 9 (15.8%) were resistant to Clindamycin, 37 (65%) to Trimethoprim sulfamexozole, 52 (91.2%) to Gentamycin and 46 (80.7%) to Ciprofloxaci 47
Conclusion: Multiple drug resistance of S. aureus isolates to antimicrobials was alarmingly high so that any empirical prophylaxis and treatment needs careful selection of effective drugs.
Keywords
MRSA; Antimicrobial agent; nosocomial; infection; staphylococcus aureus
Introduction
Staphylococcus aureus is among microbes with virulence factors that can invade immune Competent and immune-compromised human hosts. Biological adaptation and mutations have made this human pathogen a very successful infectious agent. The major characteristics of the microbe are resistance to antimicrobial agents such as methicillin, erythromycin, levofloxacin, mupirocin, and tetracycline, and its reduced susceptibility to vancomycin and daptomycin [1-3]. Both methicillin-sensitive S. aureus (MSSA) and methicillin-resistant S. aureus (MRSA) can cause mild to fatal diseases, spread locally and globally, colonize numerous human body parts. The methicillin-resistant S. aureus (MRSA) has emerged as a widespread cause of community infections and is a type of Staphylococcus that is resistant to certain antibiotics, such as methicillin, cloxacillin, dicloxacillin, oxacillin, nafcillin, and closely related class of drugs, such as cephalosporin’s (e.g. cephalexin) [2, 3]. The first report of methicillin resistance was identified in 1961. The specific gene responsible for methicillin resistance was not identified until over 20 years later [2].
The structural gene for methicillin resistance, mecA, encodes a novel penicillin-binding protein (PBP)-2a (or PBP2`), which has reduced affinity for β-lactam antibiotics. This gene is carried on a mobile genetic element, Staphylococcal Chromosomal Cassette (SCCmec).
Data regarding the prevalence and distribution of methicillinsensitive S. aureus (MSSA) and MRSA in Africa are not much, and control measures within healthcare settings are limited due to Constraints with respect to resources and diagnostic facilities. According to a study conducted at Yekatit 12 Hospital Medical College, Addis Ababa, Ethiopia (194, 14.3 %) S. aureus was recovered out of 1360 clinical specimens analysed. Out of 194 S. aureus isolates, (34, 17.5 %) were found out to be MRSA. Ninety eight (50.5 %) S. aureus were multi drug resistant and the highest isolates were resistant to penicillin (187, 96.4 %) and least resistant for clindamycin (23, 11.9 %) and vancomycin (10, 5.1 %). MRSA strains were 100 % resistant to penicillin G, erythromycin, and trimethoprim-sulfamethoxazole and least resistant to vancomycin (10, 29.4 %) [4].
The average antibiotic susceptibility patter was 75.0% for Ampicillin, 34.4%for Chloramphenicol, 1.6% for Ciprofloxacin, 7.8% for Erythromycin, 0% for Gentamycin, 45.3% for Tetracycline and 50.0% for Co-trimoxazole according to a study done in Lacor Hospital, Uganda, on sample collected from surgical inpatients and outpatients giving an average prevalence of 53.9% for both groups of patients [5].According to a study done at Moi teaching and referral hospital Eldoret of Kenya, a total of 107 isolates of S. aureus were obtained, of which 39 (37%) were MRSA. The MRSA isolates were highly resistant to erythromycin (92%; 36/39) and tetracycline (92% 36/39) and moderately susceptible to linezolid (77% 30/39), Vancomycin (75% 29/39) and fluidic acid (67% 26/39) [6]. Another cross sectional study conducted in Asmara, Eritrea on 130 study participants, the prevalence of MRSA among the isolates was 59(72%). The isolates showed 13(15.9%) resistance to vancomycin, 9(11%) to erythromycin, and 1(1.2%) to gentamycin [7].
In the study setting, there are limited data to show the recent burden of MRSA at Tikur Anbessa Specialized Hospital. This study amid to assess the Anti-microbial susceptibility patterns of coagulase positive Staphylococcus aureus isolated from patients admitted at Tikur Anbessa specialized Hospital, Addis Ababa. The findings of the study will help to figure out Anti-microbial susceptibility patterns of coagulase positive Staphylococcus aureus so as to enhance appropriate management of Hospital acquired infection. This will result in a cost effective therapy for the patients and reduced financial burden of hospitalization.
Materials and Methods
The study was carried out at Addis Ababa University Collage of Health Sciences Black Lion Tertiary hospital, Ethiopia. It was part of a cross sectional study conducted among 413 admitted Patients admitted and suspected of HAI to assess the magnitude and risk factors of methicillin resistance Staphylococcus aureus from January 2018 to January 2019 [8]. Participants were recruited using consecutive convenient sampling technique. The hospital has 800 beds, and provides services for the community in and outside the capital as referral services such as OPD, surgery, family planning, ART, laboratory, pharmacy, ANC, Oncology, ENT, delivery and neonatal care and etc. Antibiotic susceptibility test has been done on 160 coagulase positive Aureus and 57 MRSA isolates from the total collected samples.
Laboratory Analysis
Culture and Gram staining
Clinical specimens were inoculated onto blood agar base (Oxoid, Basingstoke, Hampshire,England) to which 5 % sheep blood was added and mannitol salt agar (Oxoid, Basingstoke, Hampshire, England) by using streaking method Inoculated plates were incubated at 35-37 °c For 18 to 24 h aerobically.
Bacterial colonies showing typical characteristics of S. aureus (i.e., beta haemolytic on blood agar and colonies with golden yellow pigmentation on mannitol salt agar) were subjected to subculture on to basic media, gram stain, catalase, coagulase, and postrex staph extract. Those Catalase, coagulase, postrex and gram positive bacteria appearing in grape like cluster were considered as Staphylococcus aureus.
Antimicrobial susceptibility testing
Antimicrobial susceptibility test was carried out by Kirby Bauer disc diffusion method as per Clinical Laboratory Standards Institute (CLSI, 2017) guidelines on Muller Hinton agar (Oxoid, Basingstoke, England) for 9 anti-microbial. The growth suspension was prepared in 0.5 ml of the same broth medium and the turbidity was adjusted to match that of 0.5 McFarland standards to obtain approximately the organism number of 1 × 106 colony forming units (CFU) per ml. A sterile swab was dipped into the suspension and the excess of inoculums was removed by pressing it against the sides of the tube. Then the swab was applied to the centre of Muller Hinton agar plate and evenly spread on the medium. Antibiotic discs were placed after 15 min of Inoculation to Muller Hinton agar seeded with each isolate and were incubated for 24 h at 35-37°C.
The diameter of the zone of inhibition around the disc was measured using sliding metal caliper and interpreted as Resistant/ Intermediate/Sensitive according to Kirby-Bauer disk diffusion susceptibility test protocol of American Society for Microbiology © 2016.
Data quality Assurance
Data collection sheets were checked for their completeness, readability and clearness. Culture Results were recorded carefully and double checked before entry to statistical tool SPSS 24. All specimens were collected according to the standard operating procedure (SOP) to ensure the accuracy of data, materials and equipment used. SOPs were strictly followed verifying that media Meet expiration date and quality control (QC) parameters per CLSI standards.
Visual inspections of cracks in media or plastic Petri dishes, unequal fill, haemolyses, evidence of freezing, bubbles, and contamination were performed. QC was performed to check the quality of medium.Each new lot was quality controlled before use for testing the Staphylococcus aurous using standard strains (ATCC25923).
Statistical analysis
Data was entered and analysed using SPSS software version 24 (SPSS INC, Chicago, IL, USA).The analysis was used to see the drug susceptibility pattern of coagulase positive S. aureus and MRSA towards panels of 9 antimicrobial agents using disc diffusion assay. The descriptive statistics (mean, percentages or frequency) were calculated and used to summarize the collected data. P-values less < 0.05 was taken as statistically significant.
Results
In the determination of the susceptibility of coagulase positive S. aureus on nine selected antibiotics by disk diffusion technique, the isolates were resistant to Penicillin 142(88.8%) followed by Erythromycin 93(58.1%), Ciprofloxacin 89(55.6%), Gentamicin 87(54.4%%), Trimethoprim Sulfamethoxazole 69(43.1%), Cefoxitin 57(35.6%), ceftriaxone and Augmentin each 37(35.6%) and Clindamycin 9(5.6%), (Table 1).
| Antimicrobial agents |
Sensitive (S) |
Resistant (R) |
| Erythromycin |
67(41.9%) |
93(58.1%) |
| Cefoxitin |
103(64.4%) |
57(35.6%) |
| Clindamycin |
151(75.6%) |
9(5.6%) |
| Penicillin |
17(10.6%) |
143(89.4%) |
| Trimethoprim S. |
91(56.9%) |
69(43.1%) |
| Ceftriaxone |
103(64.4%) |
87(54.4%) |
| Augmentin |
73(45.6%) |
57(35.6%) |
| Gentamycin |
71(44.4%) |
89(55.6%) |
| Ciprofloxacin |
103(64.4%) |
57(35.6%) |
Table 1: Antibacterial Susceptibility profile of Staphylococcus aureus isolates between January 2018 and January, 2019 (n=160).
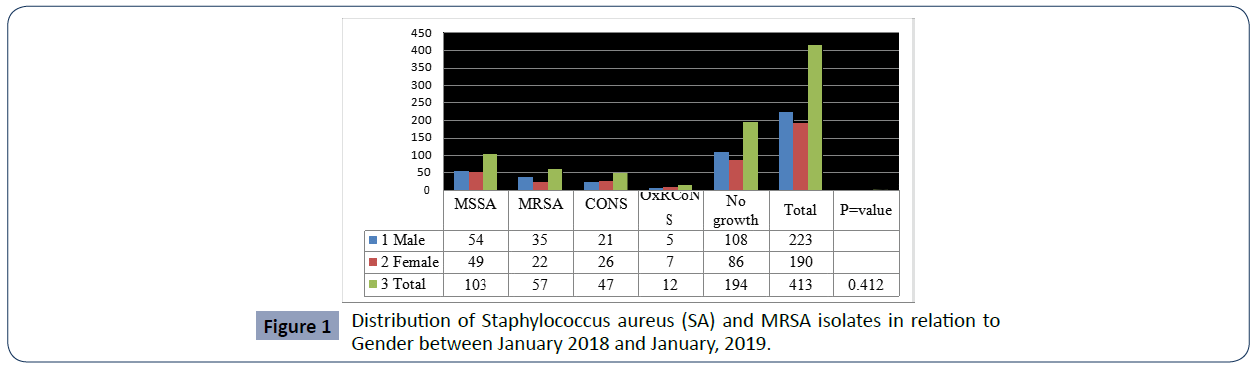
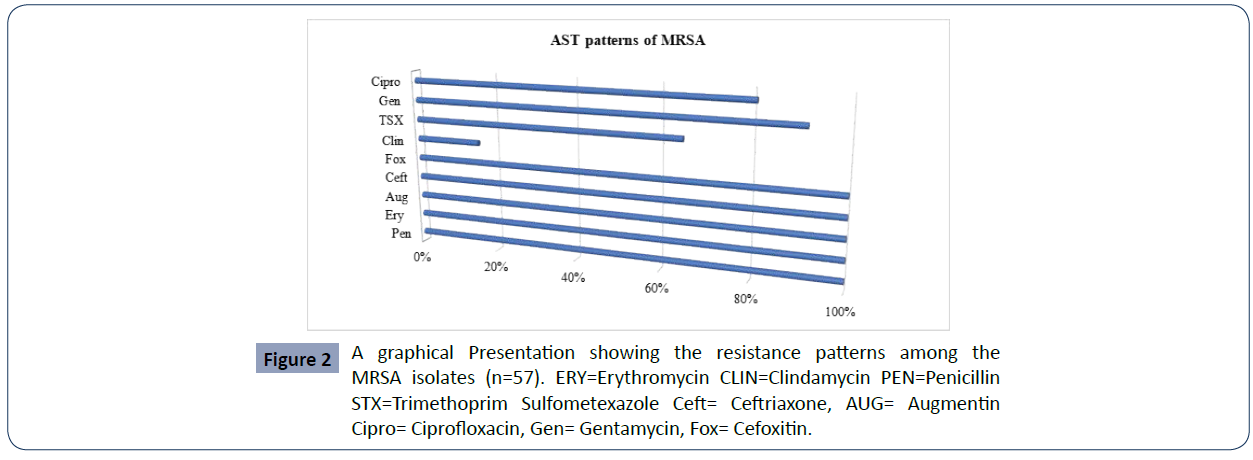
MRSA burden among the study patients was revealed to be 57/413 (13.8%), and the remaining 103/413 (25%) isolates were identified as MSSA whereas 29/413(7.02%) were CONS (Table-2). The antimicrobial susceptibility patterns of MRSA isolates were also studied. All 57 MRSA strains were 100% resistant to Penicillin, Erythromycin, Augmentin, Cefoxitin and Ceftriaxone in contrast Clindamycin (9, 15. 8 %), Trimethoprim sulfamexozole (37, 65%), Gentamycin (52, 91.2%), ciprofloxacin (46, 80.7%) resistant each respectively. (Figures 1 and 2)
Discussion
All 57 MRSA strains were 100% resistant to Penicillin, Erythromycin, Augmentin, Cefoxitin and Ceftriaxone in contrast Clindamycin (9, 15.8%), Trimethoprim sulfamexozole (37, 65%) Gentamycin (52, 91.2%), ciprofloxacin (46, 80.7%) resistant each respectively. A similar 100 % resistant to Penicillin and Erythromycin was reported in a study conducted at Yekatit 12 Hospital Addis Ababa, Ethiopia [4].
Similarity of the study population may be attributed to the resemblances of the finding. Almost comparable resistance of MRSA to Erythromycin (92%; 36/39) was reported in Kenya [6]. However, comparatively lower resistance values have been reported to Ciprofloxacin and trimethoprim-sulphamethoxazole (16.7 % each), Clindamycin (33.3 %) and erythromycin (50 %) in a study conducted at Mekelle, Northern Ethiopia [9].
High prevalence of multidrug resistant MRSA predispose patients to infection with interact And emphasizing the need for improved infection control practices and guidelines for use of antibiotics in this setting Most common reason for multi-drug resistant MRSA is indiscriminate use of antibiotics without drug sensitivity testing which may be due to lack of advanced laboratory facilities or negligence on the part of medical practitioners or patients’ poor economic status, poor hospital hygiene, and poor personal hygiene.
In Ethiopia it is a common practice that antibiotics can be purchased without prescription, which leads to misuse of antibiotics by the public, thus contributing to the Emergence and spread of antimicrobial resistance.
There was significant resistance to different antimicrobial agents by S. aureus isolates. The isolates were 88.8% (142/160) resistant to Penicillin followed by Erythromycin 58.1% (93/160), Ciprofloxacin 55.6% (89/160), Gentamicin 54.4% (87/160), Trimethoprim Sulfamethoxazole 43.1% (69/160), Cefoxitin 35.6% (5160), ceftriaxone and Augmentin each 35.6% (37/160) and Clindamycin 5.6% (9/160). Our finding is not consistent with a study finding in Lacor Hospital, Uganda (14) with average antibiotic susceptibility pattern of 7.8% for Erythromycin, and 0% for Gentamycin.
Regarding the resistance profile of isolates to individual drugs, S. aureus showed an average resistance rate of 51% to most of the antimicrobial drugs tested, which showed less resistance than the previous studies conducted in Ethiopia where average resistance of >65% were recorded [9-15]. Many factors may have contributed to the above level of resistance towards the tested antibacterial drugs, including misuse of antibiotics by health professionals, drug terminations by patients, contaminated blood transfusion, contact with pets and lack of sanitations etc.
In Our study, the overall MDR rate of S. aureus isolates among MRSA was 83.6% (resistant to three to nine drugs).Moreover all MRSA strains isolated in this investigation were resistant ≥5 antibiotics tested which was in agreement with revealed by [16]. In which almost 100% of MRSA strains were multidrug resistant. This indicated that resistant strains were emerged and the emergence of those resistant strains, especially for the most bactericidal anti- MRSA agents, may have further aggravated the emergence of multidrug resistant MRSA, and it may threaten the success of a MRSA control program [17].
Conclusion and Recommendation
It is known that antimicrobial resistance is a growing global problem. Multiple drug resistance of S. aureus isolates to antimicrobials was alarmingly high so that any empirical prophylaxis and treatment needs careful selection of effective drugs. Also there should be continuous monitoring of the antimicrobial susceptibility pattern of methicillin resistance S. aureus for the selection of appropriate therapy, developing the antibiotic policy and limiting the use of powerful antibiotics is vital. Strict consideration for S. aureus infection and proper usage of antibiotic policy are recommended in decreasing the incidence and occurrence of multidrug resistant S. aureus infections in Tikur Anbessa Specialised Hospital. It is also necessary to establish an antimicrobial susceptibility surveillance system and improve current infection control programs in the Hospital to prevent the spread of resistant MRSA.
REFERENCES
- Ildiko Roxane Bocskay (2016) Methicillin-Resistant Staphylococcus Aureus infections in the Eight Service Planning Areas of Los Angeles County Walden University.
Google Scholar, Crossref
- Nottasorn P (2012) Methicillin-Resistant Staphylococcus aureus (MRSA) Exposure Assessment in Hospital Environment. Epidemiological Science in the University of Michigan 170-181.
Google Scholar, Crossref
- Chambers HF, Deleo FR (2009) Waves of resistance: Staphylococcus aureus in the antibiotic era. Nature reviews. Microbiol 629-41.
Indexed at, Google Scholar, Crossref
- Tebelay Dilnessa, Adane Bitew (2016) Prevalence and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus isolated from clinical samples at Yekatit 12 Hospital Medical College, Addis Ababa, Ethiopia. BMC Infectious Dis 16:398.
Indexed at, Google Scholar, Crossref
- Kitara LD, Anywar AD, Acullu D, Odongo-Aginya E, Aloyo J et al (2011) Antibiotic susceptibility of Staphylococcus aureus in suppurative lesions in Lacor Hospital, Uganda. Afr Health Sci 11:S34-S39.
Indexed at, Google Scholar, Crossref
- Akoru C, Kuremu R K, Ndege S Obala A W, Smith, Bartlet M (2016) Prevalence and anti-microbial susceptibility of MRSA at Moi teaching and referral hospital Eldoret, Kenya. J Med Microbiol 6:9-16.
Indexed at, Google Scholar, Crossref
- Eyob Y, Yacob B, Oliver O, Daniel G, Robel K et al (2019) Methicillin resistance S. aureus (MRSA) prevalence and Antimicrobial Sensitivity pattern among paients. Amulti-center study in Asmara, Eritrea. Can J Infect Dis Med Microbiol Article ID:8321834.
Indexed at, Google Scholar, Crossref
- Rashedul Hasan, Mrityunjoy Acharjee, Rashed Noor (2016) Prevalence of vancomycin resistant Staphylococcus aureus (VRSA) in methicillin resistant S. Aureus (MRSA) strains isolated from burn wound infections. Department of Microbiology, Stamford University Dhaka, 297 Bangladesh; Tzu-Chi Med J 28:49-53.
Indexed at, Google Scholar, Crossref
- Goyitom G.Tewelde TG, Araya G/yesus, Tsehay A, Muthupandian S (2016) Prevalence and risk factors of methicillin-resistant Staphylococcus aureus colonization among HIV patients in Mekelle, Northern Ethiopia; Springer plus 5:877.
Indexed at, Google Scholar, Crossref
- Francesco Gesualdo (2014) Methicillin-resistant Staphylococcus aureus nasal colonization in a department of paediatrics. Ital J Pediatr 40:3.
Indexed at, Google Scholar, Crossref
- Woltering R, Hoffmann G, Daniels-Haardt I (2011) Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in patients in long-term care in hospitals, rehabilitation centers and nursing homes of a rural district in Germany; Deutsches Arzteblatt International 108:761-762.
Indexed at, Google Scholar, Crossref
- Hoganet P (2016) Prevalence of nasal colonization by methicillin-sensitive and methicillin Resistant Staphylococcus aureus among healthcare workers and students in Madagascar; BMC Infectious Dis 16:420.
Indexed at, Google Scholar, Crossref
- Katherine E, Janet P, Haas RN, Allison E, Linda RN, et al (2014) Strategies to prevent healthcare –associated infections through hand hygiene; Infection control and hospital epidemiology, USA 35:937-960.
Indexed at, Google Scholar, Crossref\
- Kitara LD, Anywar AD, Acullu D, Odongo-Aginya E, Aloyo J (2011) Antibiotic susceptibility of Staphylococcus aureus in supportive lesions in Lacor Hospital. Uganda African Health Sciences, 11:S34-S39.
Indexed at, Google Scholar, Crossref\
- Sileshi Tadesse (2014) Antimicrobial resistance profile of Staphylococcus aureus isolated from clinical specimens and nasal swabs of patients at Tikur Anbessa specialized Hospital. Addis Ababa University; 26-31.
Indexed at, Google Scholar, Crossref\
- Mulu W, Kibru G, Beyene G, Damtie M (2012) Postoperative Nosocomial Infections and Antimicrobial Resistance Pattern of Bacteria Isolates Among Patients Admitted at Felege Hiwot Referral Hospital, Bahirdar, Ethiopia; Ethiop J Health Sci 22:7-18.
Indexed at, Google Scholar, Crossref\
- Tewodros Tamire, Temesgen Eticha, Temesgen Bati Gelgelu (2021) "Methicillin-Resistant Staphylococcus aureus: The Magnitude and Risk Factors among Patients Admitted to Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia", Int J Microbiol
Indexed at, Google Scholar, Crossref
Citation: Citation: Eticha T, Tamire T, Gelgelu TB (2022) Antibiotic susceptibility pattern of coagulase positive Staphylococcus aureus isolated from patients admitted at Tikur Anbessa specialized Hospital, Addis Ababa. Health Sci J. Vol. 16 No. 2: 911