Key words
Pediocin, Pediococcus, bacteriocin, probiotic, lactic acid bacteria, cloning
Introduction
Lactic acid fermentations are deliberately exploited to produce various products such as pickled vegetables, bakery items, wine making, fermented meat, sausages and cultured milk products such as yogurts, cheeses, butter, buttermilk, kefir, koumiss etc. Natural lactic acid fermentations are brought about by lactic acid bacteria (LAB) which includes a large group of relatively fastidious, heterotrophic Gram-positive bacteria that produce lactic acid as an end product of carbohydrate fermentation. Core microbial genera of LAB include Lactobacillus, Leuconostoc, Pediococcus, Lactococcus and Streptococcus which are grouped together in the family lactobacillaceae. Their industrial importance is evidenced by their ubiquitous occurrence in natural food products, Genarally Recognized as Safe (GRAS) status, and ability to exert health benefits beyond basic nutrition. LAB display numerous antimicrobial activities which are mainly exhibited due to production of organic acids, bacteriocins and anti-fungal agents [1-6]. Highly promising results are obtained in the studies underlying the functional importance of bacteriocinogenic LAB as starter culture, consortium members and bioprotective agents in food industry that improve food quality, safety and shelf life [7]. Applications of bacteriocin starter cultures and bacteriocin thereof in various food systems are already addressed in a number of review articles [8-11]. LAB is commonly exploited in the dairy industry as producers of flavoring enzymes and metabolites that contribute to naturally rich flavor and texture of foods. A variety of probiotic LAB species including Lactobacillus acidophilus, L. bulgaricus, L. lactis, L. plantarum, L. rhamnosus, L. reuteri, L. fermentum, Bifidobacterium longum, B. breve, B. bifidum, B. esselnsis, B. lactis, B. infantis are currently recommended for development of functional food products with health-promoting capacities [12]. Health claims of various LAB strains include normalization of gastro-intestinal [13-14] and vaginal ecosystem [15-16], improvement of specific and non-specific immune responses [17], detoxification of carcinogens and suppression of tumors and cancers [18-20], reduction of blood pressure in hypertensive patients [21] and cholesterol [22]. Importance of LAB in treatment of milk allergies [23] and improvement of mineral absorption capacity of the intestine is also well documented in the literature [24].
Pediocins: The anti-microbial peptides (AMPs)
Pediococci as saprophytes were first isolated and characterized from plants by Mundt et al. [25] as catalase-negative, homofermentative bacteria producing lactic acid as a result of sugar fermentation that can tolerate temperature as high as 50°C [26]. These highly fastidious, non-motile, non-sporulating facultative anaerobes belong to family lactobacillaceae with P. acidilactici, P. pentosaceus, P. damnosus, P. parvulus, P. inopinatus, P. halophilus, P. dextrinicus, P. cellicola, P. claussenii, P. ethanolidurans and P. stilesii as the representative species. P. pentosaceus and P. acidilactici are commonly used in the fermentation of vegetables [27] and meats [28].
Anti-microbial peptides or bacteriocins are raised as an integral component of the bacterial defense mechanism and have been identified and characterized in a number of prokaryotic organisms. Bacteriocins have long attracted the interest of food sector as potential natural food preservatives against spoilage and pathogenic bacteria. Pediocins produced by various pediococcal species have gained considerable attention because of their remarkable heat stability, activity over a wide pH range, broader antimicrobial spectrum; higher specificity and effectiveness in very low concentrations [1-3, 9, 10]. A large number of pediocins have been isolated and characterized till date. Table 1 describes production of pediocins by various Pediococcal strains, class they belong to, association of their genetic determinants with small cryptic plasmids, their biochemical characteristics, mode of action and the antimicrobial spectrum.
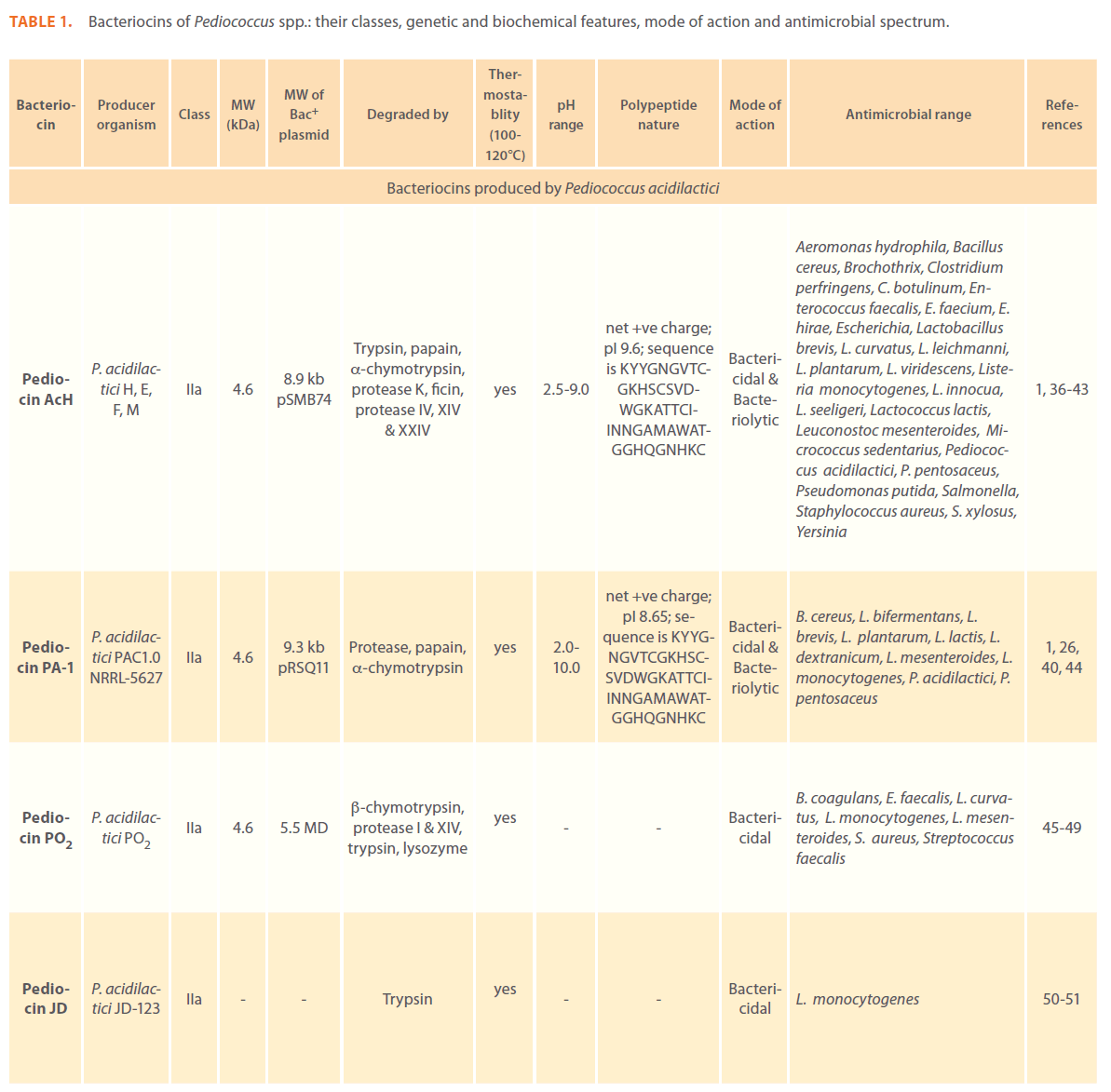
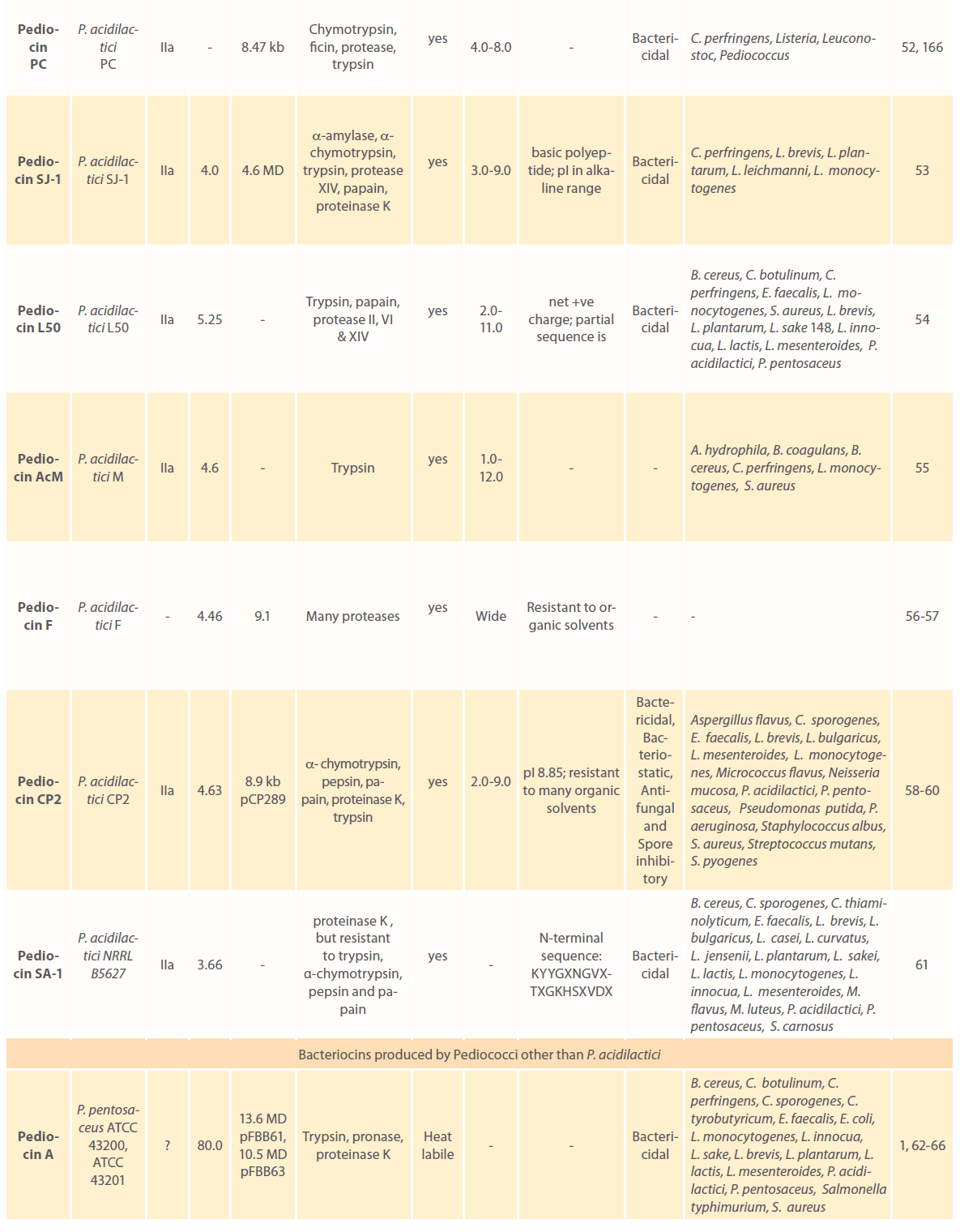
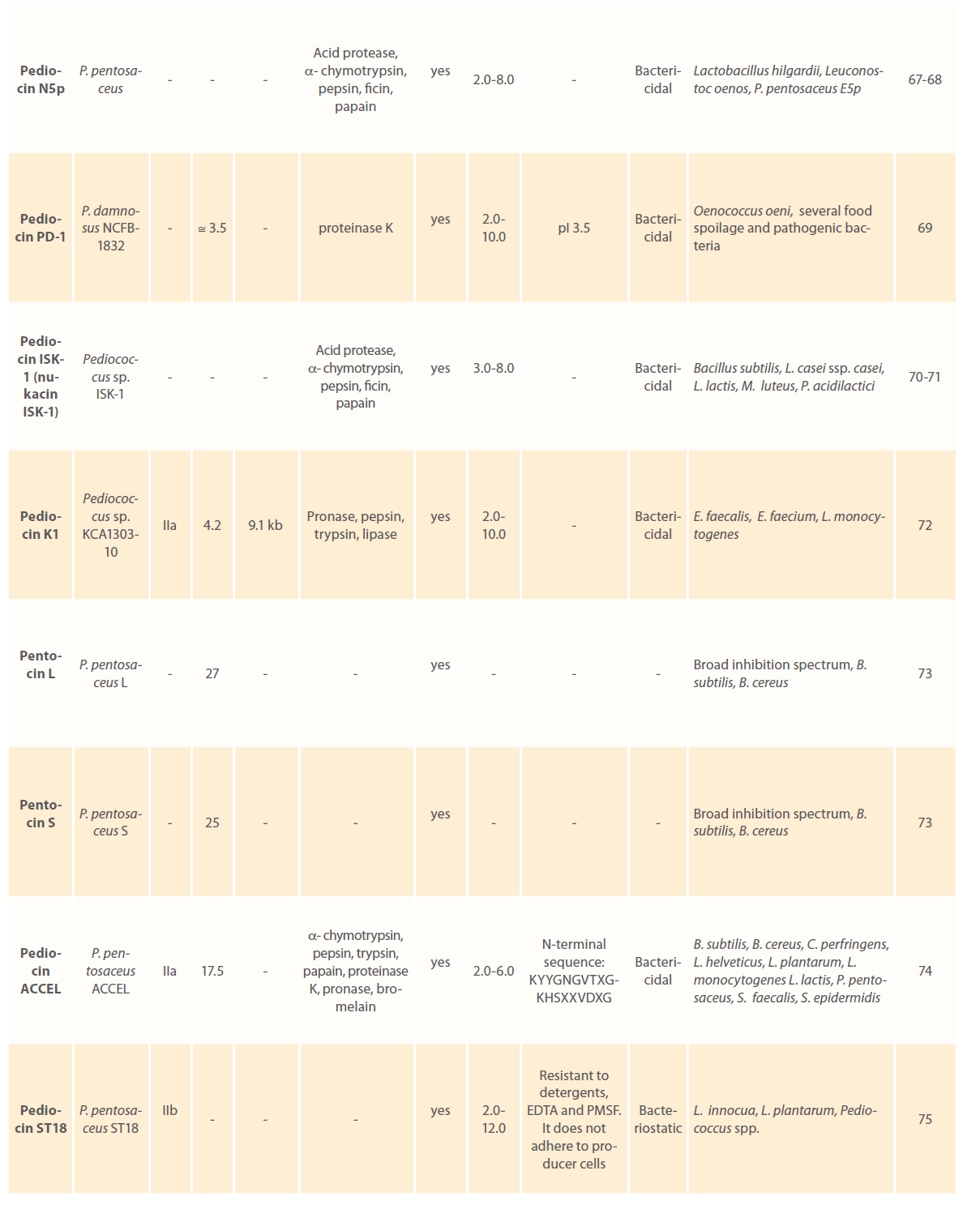
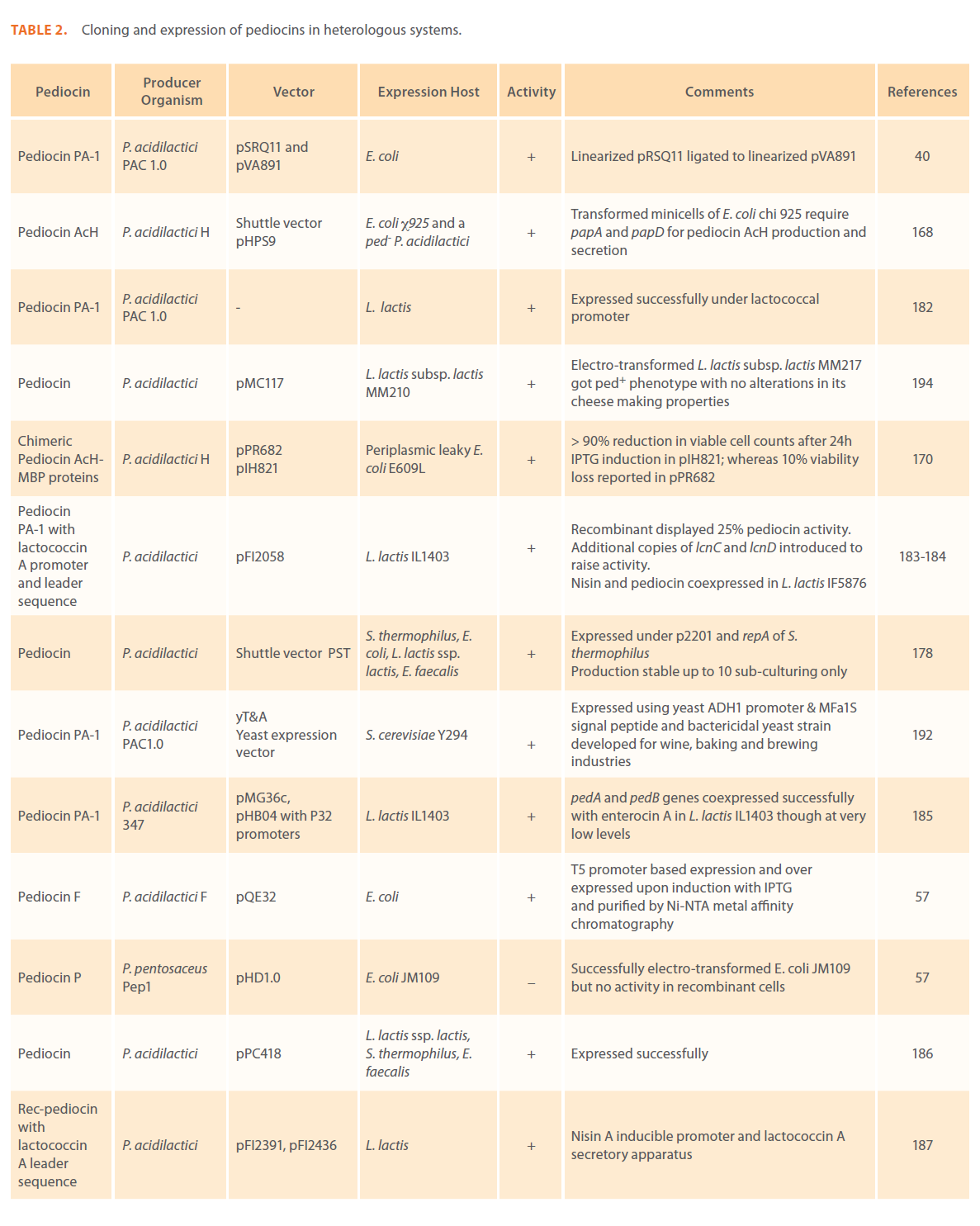
Table 1: Bacteriocins of Pediococcus spp.: their classes, genetic and biochemical features, mode of action and antimicrobial spectrum.
Classification of bacteriocins
Five classes of bacteriocins have been established based on the producing strains, common resistance mechanisms, mechanisms of action, molecular weights and chemistry [29-31]. Class I includes post-translationally modified, small lantibiotic peptides containing a number of modified amino acid residues, and it’s further divided in to two subclasses. Class Ia groups peptides with a net positive charge that exert their activity through the formation of pores in bacterial membranes (e.g. Nisin). They constitute pfam domains PF05500 and PF04369, in conjunction with F(ND)L(DEN)(LVI), SLCTPGC and SXXXCPTTXCXXXC motifs [32]. Class Ib mainly consists of post-translationally modified, small globular peptides with a negative or zero charge (e.g. Mersacidin) which’s antimicrobial activity is related to the inhibition of specific enzymes. F(ND)L(DEN)(LVI), FTCCS, GXXXTOBX-C motifs and PF05500 and PF04369 pfam domains have been identified in class Ib bacteriocins. Class IIa specifies small, strongly cationic, heat stable, non-lantibiotic, antilisterial pediocin-like peptides with at least one disulfide bridge (e.g. Pediocin PA-I, Pediocin CP2, Pediocin AcH, Enterocin A). N-terminal YGNGVXC, LSXXELXXIXGG and double glycine motifs and PF04604, PF02052, PF01721 pfam domains characterize class IIa bacteriocins. Class IIb bacteriocins require two different peptides of 25 to 65 kDa, constituting domains PF02052, PF01721 and motifs P(RQ)GXXXTOBX-C, LSXXELXXIXGG and double GG for their activity (e.g. Lactococcin G). Class IIc includes remaining cationic bacteriocins of 30 to 65 kDa which are secretory signal-dependent bacteriocins (e.g. Acidocin B). Large heat labile bacteriocins are clustered together under class III (e.g. Helveticin). Fourth class comprises an undefined mixture proteins, lipids and carbohydrates usually more than 10kDa in size. The existence of the fourth class was supported mainly by the observation that some bacteriocin activities obtained in cell free supernatant, exemplified by the activity of L. plantarum LPCO10, were abolished not only by protease treatments, but also by glycolytic and lipolytic enzymes [33]. Pediocin SJ-1, pediocin PO2 and pediocin K1 lost 50% or more acitivity upon treatment with alpha amylase, lysozyme and lipase respectively (Table 1). Thus, a situation of ambiguity arises whether to keep these heat-stable and anti-listerial bacteriocins in class IIa or class IV (Author’s own observation). One additional group of circular bacteriocins of 49-108 kDa, carrying two trans-membrane segments were housed in class V and has been described in BAGEL database [31]. BAGEL is a web-based bacteriocin genome mining tool that helps to identify putative bacteriocin ORFs in microbial genomes by extending various in silico computational methods using novel, knowledge-based bacteriocin databases and motif databases. Many bacteriocins are encoded by small genes that are often omitted in the an notation process of bacterial genomes. Gassericin A, circulatin A, and carnocyclin A are few examples of circular bacteriocins which may carry two trans-membrane segments that facilitate pore formation in sensitive cells [31, 34-35]. Their unique functional activities as well as circular nature make them potential candidates for developing novel antimicrobial agents. Class I and II bacteriocins are produced as pre-bacteriocins and usually processed during their transport through the cytoplasmic membrane at G(SA) and P(RQ) sites and GG, GG P(RQ) and G(GSA) sites respectively.
Mechanism of pediocin action
AMP’s are frequently enriched in cationic amino acid residues and interact very strongly with anionic bacterial membranes. They kill sensitive bacteria by punching holes in their cell membranes, causing a disruption in their trans-membrane potential (PMF) and destroying the delicate balance of which the organisms maintain between themselves and their environment [79]. In a study conducted on membrane vesicles derived from both sensitive and immune cells, liposome delivered pediocin PA-1 elicited efflux of small ions in a concentration dependent manner [79]. Higher concentration of pediocin effectively released, higher molecular weighted substances. They frequently adopt conformations where polar and non-polar residues are segregated properly resulting in a typical amphipathic structure that exhibits more peptide internalization and membrane perturbation. Trans-membrane potential (negative inside) in bacteria, acts as a potential driving force for insertion and internalization of the antimicrobial peptides promoting AMP interaction [80]. Pediocin PA-1 exerts bactericidal or bacteriolytic effect depending on the species of the sensitive cells [81]. Pediocins also act on some sensitive bacterial strains in bacteriostatic manner and thus retard further proliferation of the sensitive cells (e.g. Pediocin ST18, pediocin CP2). Antifungal and spore-inhibitory property of a broad spectrum pediocin CP2 has been explored in a study conducted at Department of Biotechnology, Punjabi University, Patiala, India. Antibacterial activity of bacteriocins produced by Pediococci is well documented in literature but, none of the earlier report indicates their antifungal property against A. niger isolates [82]. Currently scientists are focusing on these deadly workings of AMPs as a new approach to treat bacterial infections [12-17, 21, 83-85]. A study conducted using nisin indicated its effectiveness and efficiency as alternative therapeutic to antibiotics for the treatment of Staphylococcal mastitis [83, 84]. In vitro and in vivo studies performed with lysostaphin a class III bacteriocin have shown that this staphylococcin has potential to be used, solely or in combination with other antibacterial agents, to prevent or treat bacterial staphylococcal infectious diseases [83]. Nowadays, purified bacteriocins are available and have shown to posssess anti-neoplastic activity. Pyocin, colicin, pediocin, and microcin are among bacteriocins reported to present such activity. Moreover, modified bacteriocins proved to be effective in a glioblastoma xenograft mouse model [85].
Applications of bacteriocin producing LAB in food industry
Foodborne pathogens can multiply rapidly during extended storage at low temperature and under oxygen stress conditions, which make food unfit for consumption. Aeromonas hydrophila, Bacillus, Clostridium botulinum types B and E, Escherichia coli, Listeria monocytogenes, Pseudomonas aeruginosa, Salmonella enterinais, Shigella, Yersinia enterocolitica have been isolated from refrigerated foods and implicated in outbreaks of foodborne illness [54,86-88]. Strains of mesophilic organisms such as Salmonella and Escherichia are capable of proliferation in temperature abused (10-12°C) refrigerated systems. B. cereus has been well established as a cause of foodborne illness in humans [89-90]. Pathogenicity of B. cereus is associated with tissue destructive/ reactive exoenzyme production. It secretes a proteinaceous enterotoxin and induces a diarrheal syndrome. In addition to food poisoning, it causes a number of systemic and local infections in both immunologically compromised and immunocompetent individuals including aplstic anemia, brain abscesses, endophthalmitis, gas gangrene, meningitis, pneumonia and pseudomembranous tracheobronchitis [91]. Many pediocins are effective in controlling growth and multiplication of such foodborne pathogens and spoilage organisms in various food systems (Table 1). Many studies have highlighted the resistance of Gram-negative species to LAB bacteriocins [89, 92]. Skytta et al. [93] reported that some selected strains of Pediococci: one of P. damnosus and two of P. pentosaceus synthesize broad spectrum bacteriocins that effectively kill Gram-negative Y. enterocolitica, P. fragi and P. fluorescens in minced meat. Increased activity of bacteriocins was observed when they were used in combination with other antagonistics factors. A few reports indicated that sublethal injury due to heating, freezing, low pH exposure, ultrahigh pressure, electroporation, presence of chemical bactericidal agents such as sodium acetate, detergents and chelating agents enhance susceptibility of Gram-negative bacteria such as A. hydrophila, S. typhimurium, Y. enterocolitica, E. coli, P. putida, P. fluorescens etc. against LAB bacteriocins [42-43, 94-101]. The presence of bacteriocin-producing LAB could act as a potential barrier to inhibit the growth of spoilage bacteria and foodborne pathogens. Bromberg et al. [102] tested 813 strains of LAB which were able to inhibit the growth of Staphylococcus aureus CTC33 and/ or Listeria innocua Lin11 invitro in meat and meat products against a range of Gram-positive (B. cereus, C. sporogenes, C. perfringens, E. faecalis, L. plantarum, S. aureus) and Gram-negative (E. coli, Pseudomonas sp., S. typhimurium) test organisms and found that, Of these 128 strains showed various inhibition frequencies.
Today consumers’ preference for safe, fresh-tasting, ready-toeat, minimally-processed foods has created the necessity of exploration of novel and natural alternatives to chemical preservatives, which are useful to control development of food spoilage and pathogenic microorganisms in food systems. Nisin is a good example of food bio-preservative as well as an additional hurdle factor for increasing the shelf-life of minimal processed foods [103]. Antimicrobial substances produced by LAB offer potential applications in food preservation, food safety as well as to develop “novel” foods, health care, and pharmaceutical products [9, 88]. Bacteriocins could be added to canned/packaged food items in the form of concentrated preparations, or they could be produced in situ by bacteriocin producing starter cultures. Immobilized bacteriocins could be exploited to develop bioactive food packaging materials. Foods are considered as highly complex ecosystems where microbial interactions may influence bacteriocin efficacy and proliferation of harmful bacteria. There is a necessity to understand the global effects of bacteriocins in food ecosystems, to study bacterial genomes which may reveal new sources of bacteriocins and to develop genetically engineered food grade expression systems for development of commercial products.
Therapeutic potential of LAB bacteriocins
In the past 4 to 5 decades, use of antibiotics to fight against infectious diseases caused by microorganisms, has lead to dramatic rise of average life expectancy in humans. Unfortunately, the eventual appearance of strains resistant to multiple antibiotics in disease-causing microbes is an increasing public health problem in recent years. Urogenital problems such as bacterial vaginosis, gastrointestinal infections, pneumonia, septicemia and childhood ear infections are just a few of such diseases that have become hard to treat with antibiotics. Very often, bacteria develop several ways to resist antibiotics and other antimicrobial drugs. Other factors such as poor medical facilities, increasing use and misuse of existing antibiotics in human and veterinary medicine and in agriculture has significantly worsened the problem.
Bacterial vaginosis (BV) is one such problem where an inflammation of vagina occurs when the natural microbial balance of vagina is disturbed. Gardner [104] indicated association of bacteria such as Gardnerella vaginalis, Prevotella bivia, Peptostreptococcus spp. Mycoplasma hominis, Mobiluncus and a yeast strain Candida albicans with bacterial vaginosis. BV can have adverse outcomes of pregnancy [105-112] and enhances susceptibility to infection by HIV [113], HSV type 2 [114] and other sexually transmitted diseases. Goldstein et al. [115] had demonstrated that resistance of G. vaginalis to metronidazole increased to 68% in year 2000. Recurrence rates of up to 30% within three months after treatment have been reported [116].
Helicobacter pylori infection is another problem that affects almost all patients with duodenal ulcers and 70% of cases with gastric ulcers [117]. Pathogen weakens the protective mucous coating of the stomach and duodenum by secreting urease, protease or phospholipases etc. as virulence traits helping colonization of the pathogen. Both acid and bacteria irritate the lining and cause a sore, or ulcer [118]. Peptic ulcers are usually treated by antibiotics, proton pump inhibitors, antiacids and H2 blockers [12, 119-120]. However, emergence of antibiotic resistance in H. pylori due to point mutations and decreased binding of the antibiotics to the ribosomes has raised the concern [121-125].
Lactobacillus paracasei CRL1289 shows strong inhibition of S. aureus induced urinogenital infection as tested in a mouse model [126]. Probiotic LAB provides best alternative and attractive proposition to get rid of these opportunistic pathogens of vaginal and gastrointestinal tract. Skarin and Sylwan [127] studied growth inhibitory properties of vaginal lactobacilli against bacterial species associated with BV. Lactacin A164 produced by L. lactis subsp. A164, lacticin BH5 produced by L. lactic subsp. BH5, bulgaricin BB18 produced by L. bulgaricus BB18 and enterocin MH3 produced by Enterococcus faecium MH3 have shown strong anti-Helicobacter pylori activity in laboratory experiments [128,129]. Thus, bacteriocin producing starter cultures are potential candidates for formulating health promoting functional food products or vaginal creams which might be used to contribute a beneficial effect on the balance of intestinal and vaginal microflora respectively.
Probiotics: Best alternative to antibiotic therapy
Prof. Metchnikoff [130] the Nobel laureate of 1908, introduced the concept of probiotics in his book “The Prolongation of Life”. He argued that these friendly living bacteria normalize bowel habits, fight against disease-carrying bacteria and extend normal life span. Term “Probiotic” was first introduced by Kollath [131]. Fuller [132] gave a widely accepted definition of probiotics as “A live microbial feed supplement which beneficially affects the host animal by improving its intestinal microbial balance”. Since their establishment in various food systems, they are widely recommended in rotavirus diarrhea, to get rid of antibiotic- associated side effects, food allergies, lactose intolerance, atopic eczema, irritable bowel syndrome, inflammatory bowel disease, cystic fibrosis, traveller’s diarrhea, dental caries, to enhance proficiency of oral vaccines and to reduce incidence of certain cancers [12, 133]. The protective role of probiotics was established in colon [20, 134] and cervical cancer [135].
The indigenous microbiota plays an important role in protecting the host from colonization by opportunistic pathogens. Earlier studies have highlighted the inhibitory affects of the LAB towards BV associated pathogens [127, 136]. Lactobacillus is the predominant genus in the vaginal and endocervical microbial communities [137-139]. A number of studies explored the role of bacteriocin-like substances from vaginal isolates of Lactobacillus sps. to inhibit the growth of vaginal pathogens like E. coli, E. faecalis, E. faecium, G. vaginalis, Klebsiella spp., N. gonorrhoeae, S. aureus and Streptococcus agalactiae [140-142]. Lactocin 160, a bacteriocin produced by a probiotic vaginal L. rhamnosus has been shown to target cytoplasmic membrane of G. vaginalis [143]. The potential use of human lactobacilli as probiotics assigned to restore and maintain a healthy urogenital tract represents a promising alternative to conventional chemotherapy [12, 144-161].
Pediocin production: A plasmid linked trait in pediococci
In last two decades, there have been significant advances in functional genomic analyzes of LAB and biochemical characterization of bacteriocins produced by them. Considerable efforts have been made to functionally characterize bacteriocin operons and to express them in heterologous systems [57, 170, 178-195]. Whole genome sequence and sequences of several cryptic plasmids of Pediococci bearing genetic determinants for bacteriocin production can be retrieved from GenBank database of NCBI. As far as genetic characterization is concerned, pediocin PA-1 produced by various P. acidilactici strains has been studied extensively. Gonzalez and Kunka [26] showed that pediocin PA-1 operon of P. acidilactici PAC1.0 NRRL-5627 is located on 9.3 kb plasmid pRSQ11. Bhunia et al. [36] isolated a bacteriocin producing strain P. acidilactici H from fermented sausage. Subsequently, in their laboratory, they also identified three more Bac+ strains; E, F and M, from different sources capable of producing pediocin AcH. Pediocin production trait in all of these strains has been linked to 8.9 kb plasmid pSMB74 [37, 162-164]. P. acidilactici strains harbour this high copy number plasmid which is generally lost from the cells under stress [1] and could be transferred to plasmidless P. acidilactici strains [163]. Plasmid pSMB74 has been completely sequenced, mapped and fragments have been cloned in a pUC119 vector [165]. In P. acidilactici SJ-1, only pediocin SJ-1 structural gene is associated with a 4.6 MDa plasmid, but not its immunity factor [53]. Bacteriocin production in P. acidilactici PC too is a plasmid linked feature [52, 166]. Few other reports also indicated the plasmid linkage of bacteriocin activity in Pediococcus species. Pediocins such as PO2, PC, SJ-1, L50, AcM, F, CP2, SA-1, PD-1, K1, ACCEL, SM-1, pK23-2, ST44AM, and 05-10 are other examples where association of bacteriocin production trait has been established with small cryptic plasmids [45, 52-58, 61, 69, 72, 74, 76-78]. In P. pentosaceus, production of more than 10 bacteriocins has been reported (Table 1). Pediocin A operon in P. pentosaceus FBB61 and P. pentosaceus FBB63 has been linked to plasmids of 13.6 and 10.5 MDa sizes, respectively [62, 63, 66]. Pediocin A encoding plasmid pMD136 of P. pentosaceus ATCC 43200 was characterized by restriction fragment analysis by Kantor et al. [167]. Genetic information regarding production of various bacteriocins in P. pentosaceus (N5P, PD-1, ISK-1, ACCEL, ST18, SM-1, pK23-2, 05-10, bacteriocin ST44AM and pentocins L and S) and their immunity factors is currently not available. Plasmid borne characters have a great potential for genetic manipulations and improvement of strains for conventional starter cultures used in biotechnology industry. Their ability to show antagonism against food spoilage and pathogenic microbes opens up scope for the development of food grade biopreservatives and novel therapeutics. At the same time, such plasmid encoded characters are of interest to food technologists as they could be transferred to selected strains of LAB to develop strongly competitive starter culture bacteria which are capable of predominating over natural flora by direct antagonism along with their superior fermentation characteristics.
Genetic organization of pediocin operon
Pediocin PA-1 of P. acidilactici PAC1.0 and pediocin AcH of P. acidilactici H have been shown to contain a cluster of four genes with common promoter and terminator sequences [40, 168-169]. PedA encodes a 62 amino acids long prepediocin PA-1. Eighteen residue long leader sequence from N-terminal of pre-pediocin is removed during processing and export of pediocin through producer cell membrane. Mature pediocin carries 44 amino acid residues and two intra-molecular disulphide bridges at cys9-cys14 and cys24-cys44 positions [46, 170-171]. PedB immunity gene is located downstream to pedA and encodes a protein of 112 amino acid residues. PedC a 174 amino acid long amphiphilic protein involved along with pedD protein in facilitating/accelerating the trans-membrane export of prepediocin in P. acidilactici [168]. PedD gene specifies a polypeptide of 724 amino acid residues. Deletion analysis and site specific mutagenesis of pedD resulted in complete loss of pediocin production, showing its essentiality for secretion in E. coli [40]. PedD sequence show a very high homology to members of ATP dependent transport proteins and also to a group of eukaryotic proteins involved in multidrug resistance [40]. Very high similarity of pedD was already established with HlyB, an E. coli membrane protein required for the export of hemolysin A [172]. ComA (required for competence induction in Streptococcus pneumonia) is another member of this family of ATP binding protein with high degree of similarity [40]. These proteins carry an ATP binding motif (GMSGSGKTT) [40]. Pediocin AcH is another well characterized pediocin of P. acidilactici H linked to papABCD operon involving pediocin AcH structural gene (papA), immunity function (papB), ABC transport proteins (papC and papD) that play an important role in translocation and processing of active pediocin AcH [170]. Miller and coworker [172] provided experimental evidence by random mutagenesis that all four cysteine residues in pediocin AcH are necessary for its activity, as they play a vital role in stabilization of the secondary structure of this small peptide. His-kinase and C39-protease are other genes usually found associated with bacterioicn operon and are indirectly involved in production and secretion of active bacteriocins by producer organisms [31].
Cloning and heterologous expression of pediocins
Since the establishment of pediocin production as a plasmid linked trait, studies on cloning these plasmids in heterologous systems have started. Table 2 summarizes all those efforts made to clone potentially useful pediocins and till date, a number of research groups have reviewed their sources, production, properties, genetic features, food industry applications, antimicrobial properties etc. [2, 64, 101, 173-177]. Pediocin PA-1 has been cloned and expressed in several bacterial strains including E. coli [40, 57, 168, 170, 178-181], L. lactis [178, 182-189], L. sakei [190], S.thermophilus [178, 186], E. faecalis [178, 186], P. acidilactici [168], B. longum [191], in baker’s yeast Saccharomyces cerevisiae [192] and in methylotrophic yeast Pichia pastoris [193].
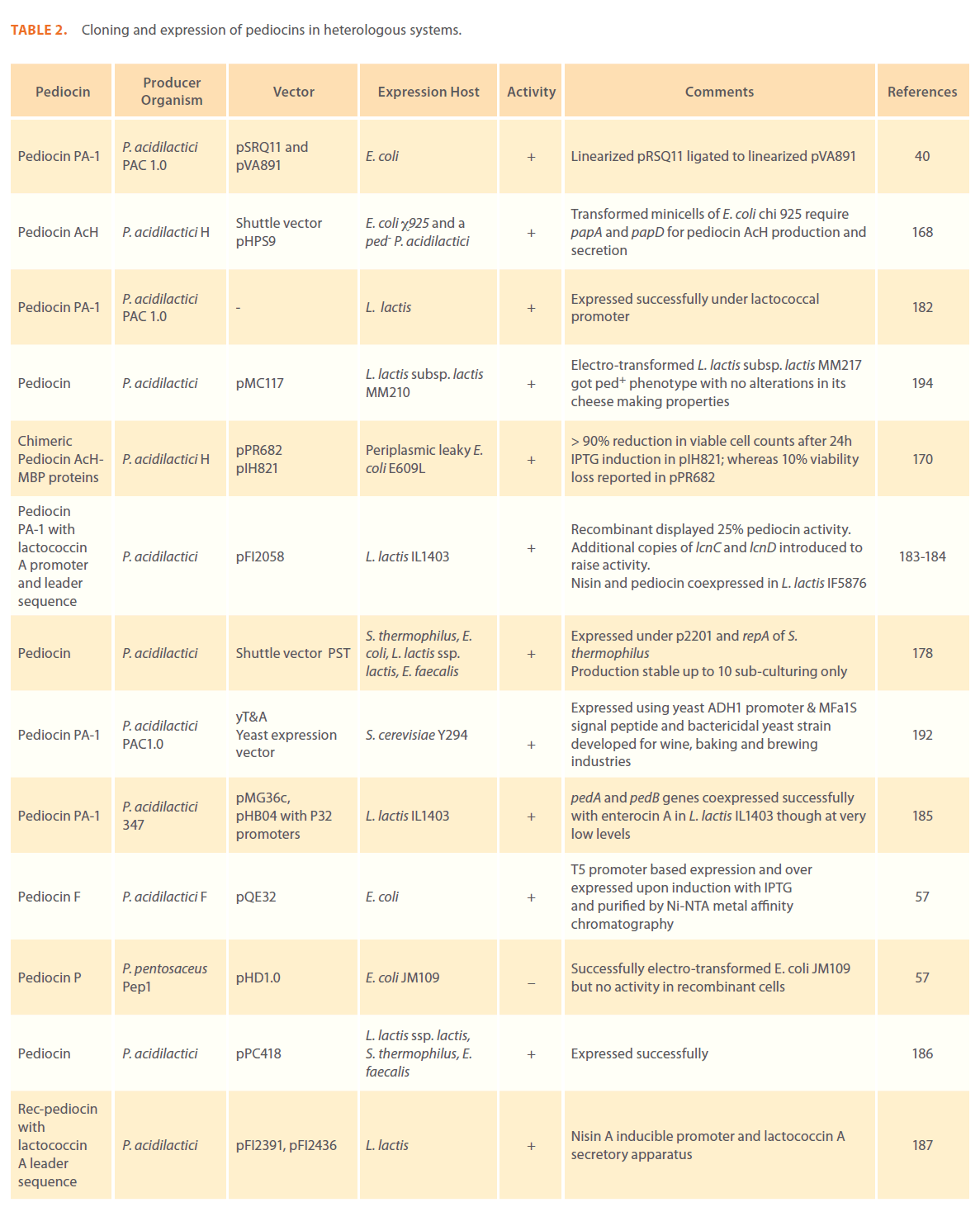
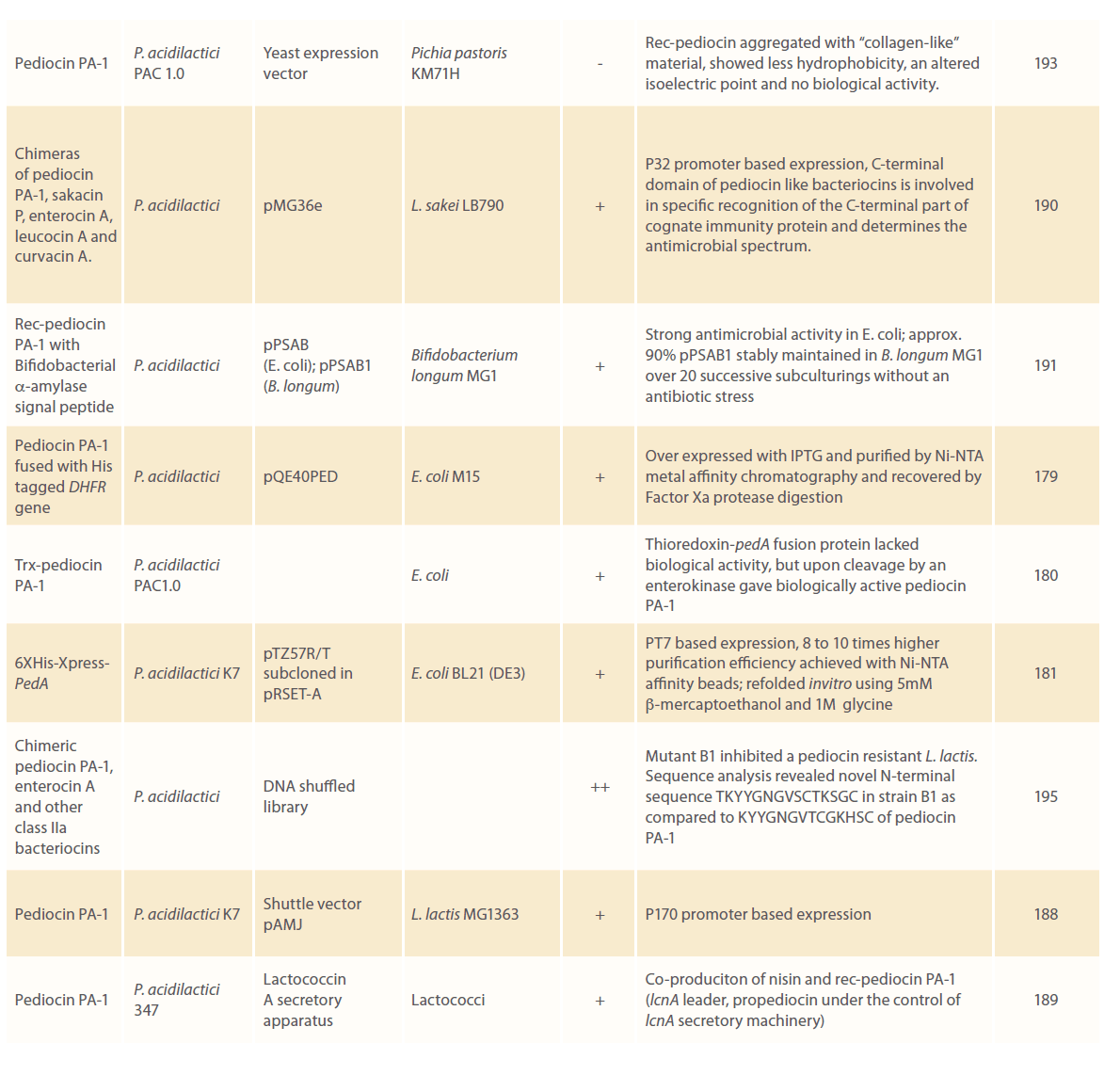
Table 2: Cloning and expression of pediocins in heterologous systems.
Cloning and expression of pediocin in E. coli
E. coli is the organism of choice for production of rec-proteins, enabling the FDA approval of Eli Lilly’s recombinant insulin under the trade name Humulin® in 1982. Afterwards the number of US and European biopharmaceutical companies grew tremendously with their ever increasing number of approved recombinant products which were cultivated in E. coli systems.
The expanding choice of E. coli expression systems for achieving high level production of rec-proteins is empowered by factors such as voluminous knowledge of their physiological and biochemical properties, availability of genetically engineered E. coli strains that facilitate formation of correct disulphide bonds in the reducing environment of cytoplasm and yield high product with least proteolytic degradation. A plethora of protease deficient E. coli strains (all B strains including B834, BL21, BLR, OrigamiTM B, RosettaTM, TunerTM are deficient in lon and ompT proteases) have been developed with their well known codon usage, as rare codons in the cloned genes can have adverse outcome on levels of protein synthesis. E. coli BL21(DE3) is most widely exploited for heterologous gene expression in E. coli. BLR(DE3) is a recA- mutant of E. coli BL21(DE3) which is commonly used to express genes carrying repetitive sequences [196]. E. coli C41(DE3) and E. coli C43(DE3) are more promising to deal with membrane proteins than native host E. coli BL21(DE3). OrigamiTM B, RosettaTM and TunerTM strains are deficient in lacY permease which facilitates uniform entry of the IPTG inducer and allows a homogenous level of induction. E. coli strains AD494, AD494(DE3), BLRtrxB, BLRtrxB(DE3), Origami, OrigamiTM B and Rosetta-gamiTM have mutations in their glutathione reductase (gor) and thioredoxin reductase (trxB) genes and have been specially designed to support formation of correct disulfide bonds in rec-proteins [197, 198]. RosettaTM are engineered to supply rare tRNA for the codons AUA, AGA, AGG, CCC, CUA and GGA on a compatible chloramphenicol resistant plasmid [199]. A very high-level expression is offered by a wide variety of tightly regulated prokaryotic promoters and expression systems (pT7Blue, pBlueStar, pRSFDuet, pSMART, pQE32, pQE40, pET32, pTZ57R/T, pRSET-A etc.). There has been a remarkable increase in the availability of fusion partners (such as T7-tag, S-tag, His-tag, HSV-tag, Trx-tag, CBD-tag, GST-tag, Nus-tag, Dsb-tag etc.) with improved protein folding tools. Recombinant proteins could be secreted by tagging with highly specific sequence tags that facilitate their detection by affinity purification, immuno-fluorescence, immuno-precipitation, western blotting. Extensive workout has been done on the mechanism of controlling gene expression and on obtaining biological activity of the proteins in heterologous E. coli systems [200].
While designing the expression systems for pediocins, one should be very particular about the natural sensitivity of the LAB against bacteriocins produced by them. Producer organisms have well developed defense machinery that protects the host from self secreted bacteriocins [40, 79, 201-204]. Thus, a need arises to co-express the pediocin immunity protein when production and secretion of the native pediocin is sought in heterologous strains. However, in some bacterial strains immunity function of pedB is not required for expression of biologically active pediocin such examples are many strains of E. coli showing resistance to pediocins produced by Gram-positive Pediococcus species [40, 168]. Shuttle vector pHPS9 bearing pedA gene from P. acidilactici H has been introduced in E. coli χ925. In transformed minicells of E. coli χ925, only papA and papD are required for pediocin AcH production and secretion, as the recombinant cells are highly resistant to pediocin AcH.
T5 promoter based expression system consisting of a Novagen vector pQE32 has been used for expression of pediocin F of P. acidilactici F in E. coli. It was over expressed upon induction with IPTG and his-tagged protein was extracted from cell lysates using Ni-NTA metal affinity chromatography [57]. Thioredoxin-pediocin PA-1 fusion protein has been expressed in E. coli. Fusion protein itself did not show any biological activity, but upon cleavage by an enterokinase, biologically active pediocin PA-1 was obtained [180]. In addition, four to five fold increases in production yield was obtained in comparison to pediocin PA-1 produced naturally by P. acidilactici PAC 1.0.
Expression of biologically active form of recombinant pediocin in non-native organisms in a soluble form remains a bottle neck. It depends upon survival tendency, propagation and copy number of recombinant plasmid in transformed host, in addition to half life of the rec-protein in an altered environment and osmotic condition of the cytosol. It has been observed that integral membrane proteins of E. coli could interfere with growth and viability of the recombinant cells, when pediocin was over expressed [170]. To overcome this problem, papA was fused in-frame to secretary maltose binding protein (MBP) of E. coli and coned in malE vectors pPR682 and pIH82, whose efficient and powerful secretory signals directed very high level synthesis of MBP chimeric protein [170]. About one third of chimeric proteins were secreted into periplasm and released into the culture medium by periplasmic leaky E. coli E609L. However, a very high viability loss of >90% in recombinant E. coli E609L transformed with pIH821 and of 10% in E. coli E609L transformed with pPR682 was observed after 24h of IPTG induction.
Upon over-expression in heterologous systems, rec-proteins may tend to accumulate in inclusion bodies (IBs) of E. coli as a result of reducing conditions of the cytosol. To extract an intracellular protein it is necessary to disrupt the cells and separate IBs [205]. IBs are subsequently washed and resolublized for proper folding of rec-proteins [206]. 6XHis-Xpress-pedA carrying pediocin structural gene from P. acidilactici K7 was cloned in pTZ57R/T [181]. It was further subcloned in pRSET-A for over expression in E. coli BL21(DE3). Recombinant pediocin was purified using Ni-NTA beads and eluted with 0.5M imidazole. In vitro refolding of rec-pediocin was carried out in redox system consisting of 5mM b-mercaptoethanol and 1M glycine to achieve its biological activity.
The antimicrobial activity of the heterologous expressed pediocin varied from 0 to 10 fold depending on the expression system used. Osmanagaoglu et al. [57] successfully electrotransformed E. coli JM109 cells with pHD1.0 bearing pediocin P structural gene from P. pentosaceus Pep1, but none of the transformant was able to express and/or release pediocin P. To overcome this, rec-pediocin was fused inframe with alpha-amylase signal peptide of Bifidobacterium to construct plasmids pSAB and pSAB1 for transforming E. coli and B. longum MG1, respectively [191]. Recombinant E. coli showed strong antimicrobial activity, while 90% of pSAB1 was stably maintained in B. longum MG1 over 20 successive subculturings without an antibiotic stress. Moon and coworkers [179] fused pedA with His-tagged DHFR in pQE40PED and transformed E. coli M15. Recombinants displayed very high pediocin activity upon overexpression with IPTG and subsequently fusion protein was purified by Ni-NTA affinity chromatography. Recovery of the native pediocin PA-1 from fusion product was achieved by digestion with Factor Xa protease. PT7 based expression system offers 8 to 10 times higher yields with great purification efficiency achieved through Ni-NTA affinity beads [181].
Cloning and expression of pediocin in other microbial systems
Heterologous hosts including S. thermophilus, L. lactis subsp. lactis and E. faecalis have been demonstrated for their ability to express pediocin under p2201 and repA of shuttle vector PST [178]. The major limitation of these expression systems is the decreased stability (upto 10 subculturings only) of the cloned genes. A chimeric stretch consisting of lactococcin A promoter, lactococcin A leader sequence and pediocin PA-1 structural gene has been introduced in pFI2058 for constructing a recombinant plasmid which was used to transform L. lactis IL1403. Recombinant lactococcal strains displayed only 25% pediocin activity. Thus, in an attempt to raise pediocin yields, additional copies of lcnC and lcnD were co-introduced in recombinant L. lactis IL1403. Using same recombinant pFI2058, a nisin producing L. lactis IF5876 was also transformed, where nisin and pediocin PA-1 were coexpressed successfully [183- 184]. PedA and pedB genes of pediocin operon from P. acidilactici 347 have been successfully coexpressed with enterocin A in L. lactis IL1403 using plasmids pMG36c, pHB04 carrying P32 promoters, but resulting pediocin activity detected in recombinant cells was very low [185]. Rec-pediocin with lactococcin A leader sequence was secreted by recombinant L. lactis bearing plasmids pFI2391, pFI2436 under nisin inducible promoters and lactococcin A secretory apparatus [187]. P170 promoter based expression system has also been exploited for over-expression of rec-pediocin in L. lactis MG1363 using the shuttle vector pAMJ [188].
DNA shuffling technique has enabled construction of chimeric gene sequences carrying desirable traits. Chimeras of pediocin PA-1, sakacin P, enterocin A, leucocin A and curvacin A were generated by shuffling the genes of five different parental bacteriocins. Subsequent cloning of chimeric constructs in P32 promoter based expression vector pMG36e was accomplished and recombinant L. sakei LB790 was generated [190]. Results indicated that some of the variants have dramatically more bacteriocin activity than their native bacteriocins. Results also highlighted the involvement of C-terminal domain of pediocin like bacteriocins in specific recognition of the cognate immunity protein and determination of the antimicrobial spectrum of the secreted bacteriocin.
Attempts have been made to express pediocin in yeast strains S. cerevisiae and P. pastoris, where active disulphide bond formation can take place; however studies showed low levels of expression [192] and inhibition of its biological activity [193]. Aggregation of the rec-pediocin was observed in P. pastoris KM71H, due to its association with collagen-like material. These collagen-pediocin aggregates were less hydrophobic and behaved differently when subjected to isoelectric focusing. Recpediocin lost its biological activity due to aggregation [193].
Conclusions
Though pediocin is an equally promising biopreservative as nisin is, its indusrial scale production has not been taken up yet. The main reason is lack of a comparable scale of production. To improve its production heterologous systems have been studied which have used a variety of promoters for enhanced expression, secretory proteins for fusion and peptide tags to facilitate purification. Present review compiled the information available to date, giving variety of production enhancing strategies for improving heterologous pediocin production. Apart from its biopreservative potential in foods, pediocin is an attractive antimicrobial agent against many pathogenic bacteria and hence has pharmaceutic application too. As an additive to cosmetics its property to modulate skin microflora needs to be explored. Its probiotic potential in modulating gut microbiota towards cholesterol lowering, antidiabetic and antihypertensive state promises to make it an important component of neutraceutic and wellness products. For all these applications either GRAS grade whole cells, over secreting copious amounts of pediocin or purified pediocin producted at industrial scale can be used. More research into production aspects is needed in near future.
216
References
- Ray B and Daeschel MA (1994) In: Dillon VM, Board RG, editors. Natural antimicrobial systems and food preservation. CAB International, Wallingford, Oxon, UK, pp. 133–165.
- Jack RW, Tagg JR and Ray B (1995) Bacteriocins of Gram positive bacteria. Microbiol Rev 59: 171–200.
- Lavermicocca P, Valerio F, Evidente A, Lazzaroni S, Corsetti A, et al. (2000) Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Appl Environ Microbiol 66: 4084–4090.
- Magnusson J and Schnürer J (2001) Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad spectrum proteinaceous antifungal compound. Appl Environ Microbiol 67: 1–5.
- Schnürer J and Magnusson J (2005) Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci Technol 16: 70–78.
- Prema P, Smila D, Palavesam A, Immanuel G (2008) Production and characterization of an antifungal compound (3–Phenyllactic acid) produced by Lactobacillus plantarum strain. Food Bioprocess Technol 3: 379–386.
- Liu M, Bayjanov JR, Renckens B, Nauta A, Siezen RJ (2010) The proteolytic system of lactic acid bacteria revisited: a genomic comparison. BMC Genomics 11: 36. doi-10-1186/147-2164-11-36.
- Bhardwaj A, Malik RK and Chauhan P (2008) Functional and safety aspects of enterococci in dairy foods. Ind J Microbiol 48(3): 317–325.
- Gálvez A, Abriouel H, López RL, Omar NB (2007) Bacteriocin– based strategies for food biopreservation. Int J Food Microbiol 120: 51–70.
- Vuyst LD and Leroy F (2007) Bacteriocins from lactic acid bacteria: production, purification and food applications. J Mol Microbiol Biotechnol 13: 194–199.
- Olaoye OA and Onilude AA (2009) A study on isolation of presumptive technologically important microorganisms from nigerian beef. Am–Eurasian J Sustain Agric 3(1): 75–83.
- Kaur B, Balgir PP, Kumar B, Garg N (2010) Helicobacter pylori infection: efficacy of probiotics and role of genome wide association studies. Arch Clinical Microbiol 1(4). doi: 10:3823/216.
- Szajewska H, Setty M, Mrukowicz J, et al. (2005) Probiotics in gastrointestinal diseases in children: hard and not–so–hard evidence of efficacy. J Pediatr Gastroenterol Nutr 42: 454–475.
- McFarland LV (2007) Meta–analysis of probiotics for the prevention of traveler’s diarrhea. Travel Med Infect Dis 5(2): 97–105.
- Reid G, Beuerman D, Heinemann C, Bruce AW (2001) Probiotic Lactobacillus dose required to restore and maintain a normal vaginal flora. FEMS Immunol Med Microbiol 32: 37–41.
- Falagas ME, Betsi GI and Tokas T (2006) Probiotic for the prevention of recurrent UTI in women. J Microbes Infect 12: 2772–2776.
- Schiffrin EJ, Brassart D, Serving AL, Rochat F, Donnet–Hughes A (1997) Immune modulation of blood leukocytes in human by lactic acid bacteria: criteria for strain selection. Am J Nutrition 66(2): 515S–520S.
- Kulkarni N and Reddy BS (1994) Inhibitory effect of B. longum cultures on the azoxymethane induced aberrant crypt foci formation on focal bacterial beta–gluconidase. Proc Soc Exp Biomed 207: 278–283.
- Sekine K, Watanabe–Sekine E, Ohta J, et al. (1994) Induction and activation of tumoricidal cells in vitro and in vivo by the bacterial cell wall of B. infantis. Bifidobacteria Microflora 13: 54–77.
- Wollowski I, Rechkemmer G and Pool–Zobel BL (2001) Protective role of probiotics and prebiotics in colon cancer. Am J Clin Nutr 73(2): 451–455.
- Sanders ME (2000) Considerations for use of probiotic bacteria to modulate human health. J Nutr 130(2): 384–390.
- Simons LA, Amansec SG and Conway P (2006) Effect of Lactobacillus fermentum on serum lipids in subjects with elevated serum cholesterol. Nutr Metab Cardiovas Dis 16: 531– 535.
- Kirjavainen PV, Salminen SJ and Isolauri E (2003) Probiotic bacteria in the management of atopic disease: underscoring the importance of viability. J Pediatr Gastroenterol Nutr 36(2): 223–227.
- Famularo G, Minisola G, Nicotra GC, De Simone C (2005) Acute pancreatitis associated with irbesartan therapy. Pancreas 31: 294–295.
- Mundt JO, Bealtie WG and Wieland FR (1969) Pediococci residing on plants. J Bacteriol 98: 938–942.
- Gonzalez CF and Kunka BS (1987) Plasmid– associated bacteriocin production and sucrose fermentation in Pediococcus acidilactici. Appl Environ Microbiol 53: 2534–2538.
- Smith JL and Palumbo SA (1983) Use of starter cultures in meats. J Food Prot 46: 997–1006.
- Nes IF, Bao Diep D, Havarstein LS, Brurberg MB, Eijsink V, et al. (1996) Biosynthesis of bacteriocins of lactic acid bacteria. Antonie van Leeuwenhoek 70: 113–128.
- Kawai Y, Ishii Y, Arakawa K, Uemura K, Saiton B, et al. (2004) Structural and functional differences in two cyclic bacteriocins with the same sequences produced by Lactobacilli. Appl Environ Microbiol 70(5): 2906–2911.
- Jong AD, van Hijum SAFT, Bijlsma JJE, Kok J, Kuipers OP (2006) BAGEL: a web–based bacteriocin genome mining tool. Nucleic Acids Res 34(2): 273–279.
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer ELL, Eddy SR, Bateman A (2010) The Pfam protein families database. Nucleic Acids Res 38: 211-222.
- Jimenez–Diaz R, Rios–Sanchez RM, Desmazeaud M, Ruiz–Barba JL, Piard JC (1993) Plantaricin S and T, two new bacteriocins produced by Lactobacillus plantarum LPCO10 isolated from a green olive fermentation. Appl Environ Microbiol 59: 1416–1424.
- Kawai Y, Kemperman R, Kok J, Saito T (2004) The circular bacteriocins gassericin A and circularin A. Curr Protein Pept Sci 5: 393–398.
- Gong X, Martin–Visscher LA, Nahirney D, Vederas JC, Duszyk M (2009) The circular bacteriocin, carnocyclin A, forms anion– selective channels in lipid bilayers. Biochem Biophys Acta 1788(9): 1797–1803.
- Bhunia AK, Johnson MC and Ray B (1987) Direct detection of an antimicrobial peptide of Pediococcus acidilactici in SDS–PAGE. J Indust Microbiol 2: 319–322.
- Bhunia AK, Johnson MC and Ray B (1988) Purification, characterization and antimicrobial spectrum of a bacteriocin produced by Pediococcus acidilactici. J Appl Bacteriol 65: 261– 268.
- Bhunia AK, Johnson MC, Ray B, Belden EL (1990) Antigenic property of pediocin AcH produced by Pediococcus acidilactici H. J Appl Bacteriol 69: 211–215.
- Marugg JD (1991) Bacteriocins, their role in developing natural products. Food Biotechnol 5: 305–312.
- Marugg JD, Gonzalez CF, Kunka BS, Ledeboer AM, Pucci MJ, et al. (1992) Cloning, expression and nucleotide sequence of genes involved in production of pediocin PA–1, a bacteriocin from Pediococcus acidilactici PAC1.0 . Appl Environ Microbiol 58: 2360– 2367.
- Ennahar S, Aoude–Werner D, Sorokine O, Van Dorsselaer A, Oise bringel F, et al. (1996) Production of pediocin AcH by Lactobacillus plantarum WHE92 isolated from Cheese. Appl Environ Microbiol 62(12): 4381–4387.
- Kalchayanand N, Hanlin MB and Ray B (1992) Sublethal injury makes gram–negative and resistant gram–positive bacteria sensitive to the bacteriocin, pediocin AcH and nisin. Lett Appl Microbiol 15: 239–243.
- Kalchayanand N, Sikes T, Dunne CP, Ray B (1994) Hydrostatic pressure and electroporation have increased bactericidal efficiency in combination with bacteriocins. Appl Environ Microbiol 60: 4174–4177.
- Pucci MJ, Vedamuthu ER, Kunka BS, Vandenbergh PA (1988) Inhibition of Listeria monocytogenes by using bacteriocin PA–1 produced by Pediococcus acidilactici PAC1.0. Appl Environ Microbiol 54: 2349–2353.
- Hoover DG, Walsh PM, Kolaetis KM, Daly MM (1988) A bacteriocin produced by Pediococcus species associated with a 5.5– megadalton plasmid. J Food Prot 51: 29–31.
- Nieto–Lozano JCN, Nissen–Meyer J, Sletten K, Pela´z C, Nes IF (1992) Purification and amino acid sequence of a bacteriocin produced by Pediococcus acidilactici. J Gen Microbiol 138: 1985– 1990.
- Liao CC, Yousef AE, Richter R, Chism GW (1993) Pediococcus acidilactici PO2, bacteriocin production in whey permeate and inhibition of Listeria monocytogenes in foods. J Food Sci 58: 430–434.
- Coventry MJ, Gordon JB, Alexander M, Hickey MW, Wan J (1996) A food–grade process for isolation and partial purification of bacteriocins of lactic acid bacteria that uses diatomite calcium silicate. Appl Environ Microbiol 62: 1764–1769.
- Cho HY, Yousef AE and Yang ST (1996) Continuous production of pediocin by immobilized Pediococcus acidilactici PO2 in a packed–bed bioreactor. Appl Environ Microbiol 62: 589–594.
- Berry ED, Liewen MB, Mandigo RW, Hutkins RW (1990) Inhibition of Listeria monocytogenes by bacteriocin producing Pediococcus during the manufacture of fermented semidry sausage. J Food Prot 53: 194–197.
- Berry ED, Hutkins RW and Mandigo RW (1991) The use of bacteriocin producing Pediococcus acidilactici to control post processing Listeria monocytogenes contamination of frankfurters. J Food Prot 54: 681–686.
- Jager K and Harlander S (1992) Characterization of a bacteriocin from Pediococcus acidilactici PC and comparison of bacteriocin– producing strains using molecular typing procedure. Appl Microbiol Biotechnol 37: 631–637.
- Schved F, Lalazar A, Henis Y, Juven BJ (1993) Purification, partial characterization and plasmid–linkage of pediocin SJ–1, a bacteriocin produced by Pediococcus acidilatici. J Appl Bacteriol 74: 67–77.
- Cintas LM, Rodriguez JM, Fernandez MF, Sletten K, Nes IF, et al. (1995) Isolation and characterization of pediocin L50, a new bacteriocin from Pediococcus acidilactici with a broad inhibitory range. Appl Environ Microbiol 61: 2643–2648.
- Elegado FB, Kim WJ and Kwon DY (1997) Rapid purification, partial characterization and antimicrobial spectrum of the bacteriocin, pediocin AcM, from Pediococcus acidilactici M. Int J Food Microbiol 37: 1–11.
- Osmanagaoglou O, Gunduz U, Beyatli Y, Cokmus C (1998) Purification and characterization of pediocin F, a bactericin produced by Pediococcus acidilactici F. Tr J Biol 22: 217–228.
- Osmanagaoglu O, Beyatli Y, Gündüz U (2000) Cloning and expression of a plasmid–linked pediocin determinant trait of Pediococcus acidilactici F. J Basic Microbiol 40(1): 41–49.
- Kaur B and Balgir PP (2007) Pediocin CP2 gene localization to plasmid pCP289 of Pediococcus acidilactici MTCC 5101. Internet J Microbiol 3(2).
- Kaur B and Balgir PP (2008) Biopreservative Potential of a broad– range pediocin CP2 Obtained from Pediococcus acidilactici MTCC 5101. Asian J Microbiol Biotechnol Environ Sci 10(2): 439–444.
- Kaur B, Kumar B, Balgir PP, Bhatia P (2009) Comparative evaluation of media for pediocin production production by Pediococcus acidilactici CP2 isolate. Int J Probiot Prebiot 4(4): 233–240.
- Anastasiadou S, Papagianni M, Filiousis G, Ambrosiadis I, Koidis P (2008) Pediocin SA–1, an antimicrobial peptide from Pediococcus acidilactici NRRL B5627: production conditions, purification and characterization. Biores Technol 99: 5384–5390.
- Daeschel MA and Klaenhammer TR (1985) Association of a 13.6 M Dal plasmid in Pediococcus pentosaceus with bacteriocin activity. Appl Environ Microbiol 50: 1538–1541.
- Grahm DC and McKay LL (1985) Plasmid DNA in strains of Pediococcus cerevisiae and Pediococcus pentosaceus. Appl Environ Microbiol 50: 532–534.
- Klaenhammer TR (1988) Bacteriocins of lactic acid bacteria. Biochem 70: 337–349.
- Skytta E, Hairkara A and Sandholm TM (1993) Production and characterization of antibacterial compounds produced by Pediococcus damnosus and P. pentosaceus. J Appl Bacteriol 74: 134–142.
- Piva A and Headon DR (1994) Pediocin A, a bacteriocin produced by Pediococcus pentosaceus FBB61. Microbiol 140: 697–702.
- Strasser de Saad AM and Manca de Nadra MC (1993) Characterization of bacteriocin produced by Pediococcus pentosaceus from wine. J Appl Bacteriol 74: 406–410.
- Strasser de Saad AM, Pasteris SE and Manca de Nadra MC (1995) Production and stability of pediocin N5p in grape juice medium. J Appl Bacteriol 78(5): 473–476.
- Green G, Dicks LM, Bruggeman G, Vandamme EJ, Chikindas ML (1997) Pediocin PD–1: a bactericidal antimicrobial peptide from Pediococcus damnosus NCFB1832. J Appl Microbiol 83: 127–132.
- Kimura H, Nagano R, Matsusaki H, Sonomoto K, Ishizaki A (1997) A bacteriocin of strain Pediococcus sp. ISK–I isolated from Nukadoko, bed of fermented rice bran. Biosci Biotechnol Biochem 61: 1049–1051.
- Sashihara T, Dan M, Kimura H, Matsusaki H, Sonomoto K, et al. (2000) The effect of osmotic stress on the production of nukacin ISK–1 from Staphylococcus warneri ISK–1. Appl Microbiol Biotechnol 56(3–4): 496–501.
- Kim CH, Ji GE and Ahn C (2000) Purification and molecular characterization of a bacteriocin from Pediococcus sp. KCA 1303– 10 isolated from fermented flat fish. Food Sci Biotechnol 9(4): 270–276.
- Yin LJ, Wu CW and Jiang ST (2003) Bacteriocins from Pediococcus pentosaceus L and S from pork meat. J Agr Food Chem 51: 1071– 1076.
- Wu CW, Yin LJ and Jiang ST (2004) Purification and characterization of bacteriocin from Pediococcus pentosaceus ACCEL. J Agr Food Chem 52: 1146–1151.
- Todorov SD and Dicks LMT (2005) Production of bacteriocin ST33LD, produced by Leuconostoc mesenteroides subsp.mesenteroides, as recorded in the presence of different medium components. World J Microbiol Biotechnol 21: 8–9.
- Shin MS, Han SK, Ryu JS, Kim KS, Lee WK (2008) Isolation and partial characterization of a bactericin produced by Pediococcus pentosaceus K23–2 isolated from Kimchi. J Appl Microbiol 105(2): 331–339.
- Huang Y, Luo Y, Zhai Z, Zhang H, Yang C, et al. (2009) Characterization and application of an anti–Listeria bacteriocin produced by Pediococcus pentosaceus 05–10 isolated from Sichuan Pickle, a traditionally fermented vegetable product from China. Food Control 20: 1030–1035.
- Todorov SD and Dicks LMT (2005) Pediocin ST18, an antilisterial bactericin produced by Pedicoccus pentosacceus ST18 isolated from boza, a traditional cereal beverage from Bulgaria. Process Biochem 40: 365–370.
- Chikindas ML, Garcia–Garcera MJ, Driessen AJM, Ledeboer AM, Nissen–Meyer J, et al. (1993) Pediocin PA–1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl Environ Microbiol 59: 3577–3584.
- Manuel N, Rafael M, Miguel A, Castanho RB (2009) Antimicrobial peptides: linking partition, activity and high membrane–bound concentrations. Nature Reviews Microbiol 7: 245–250.
- Bhunia AK, Johnson MC, Ray B, Kalchayanand N (1991) Mode of action of pediocin AcH from Pediococcus acidilactici H on sensitive bacterial strains. J Appl Bacteriol 70: 25–33.
- Mashal (2007) Biopermeabilization and antimicrobial applications of purified pediocin CP2 produced from P. acidilactici MTCC 5101. A project report, Department of Biotechnology, Punjabi University, Patiala, Punjab.
- Bastos MCF, Coutinho BG and Coelho MLV (2010) Lysostaphin: A Staphylococcal Bacteriolysin with Potential Clinical Applications. Pharmaceuticals 3:1139-1161.
- Fernandez L, Delqado S, Herrero H, Maldonado A, Rodriquez JM (2008) The Bacteriocin Nisin, an Effective Agent for the Treatment of Staphylococcal Mastitis During Lactation. J Hum Lact 24(3):311-316.
- Gilbert C, Claude F and Denis S (2008) Antineoplastic Properties of Bacteriocins: Revisiting Potential Active Agents. Amer J Clin Oncol 31(4): 399-404.
- Gonzalez–Fandos E, Otero A, Sierra M, Garcia–Lopez ML, Prieto M (1994) Effect of three commercial starters on growth of Staphylococcus aureus and enterotoxins (A–D) and thermonuclease production in broth. Int J Food Microbiol 24(1– 2): 21–27.
- Holzapfel WH, Geisen R and Schillinger U (1995) Biological preservation of foods with reference to protective cultures, bacteriocins and food–grade enzymes. Int J Food Microbiol 24(3): 343–362.
- Ahmed Z, Wang Y, Cheng Q, Imran M (2010) Lactobacillus acidophilus bacteriocin, from production to their application: an overview. Afr J Biotechnol 9(20): 2843–2850.
- Motlagh AM, Johnson MC and Ray B (1991) Viability loss of foodborne pathogens by starter culture metabolites. J Food Prot 54: 873–878.
- Balasubramanyam BV and Varadaraj MC (1994) Dahi as a potential source of LAB active against food borne pathogenic and spoilage bacteria. J Food Sci Technol 31: 241–243.
- Bottone EJ (2010) Bacillus cereus, a Volatile Human Pathogen. Clin Microbiol Reviews 23(2): 382–398.
- Spelhaug SR and Harlander SK (1989) Inhibition of foodborne bacterial pathogens by bacteriocins from Lactococcus lactis and Pediococcus pentosaceus. J Food Prot 52: 856–862.
- Skytta E, Hereijgers W and Mattila–Sandholm T (1991) Broad spectrum antibacterial activity of Pediococcus damnosus and Pediococcus pentosaceus in minced meat. Food Microbiol 8: 231–237.
- Blackburn P, Polak J, Gusik SA, Rubino SD (1989) Nisin composition for use as enhanced broad range bactericides. Int Patent Appl. Publ. No. WO 89112399, 1155, AVC Americus, NY, USA.
- Lyon WJ and Glatz BA (1991) Partial purification and characterization of a bacteriocin produced by Propionibacterium thoenii. Appl Environ Microbiol 57: 701–706.
- Stevens KA, Sheldon BW, Klapes NA, Klaenhammer TR (1991) Nisin treatment for the inactivation of Salmonella species and other gram–negative bacteria. Appl Environ Microbiol 57: 3613– 3615.
- Ray B (1992) In: Ray B, Daeschel MA, editors. Food biopreservatives of microbial origin. CRC Press, Boca Raton, Florida, USA, pp. 207–264.
- Ray B (1992) In: Ray B, Daeschel MA, eds. Food biopreservatives of microbial origin. CRC Press, Boca Raton, Florida, USA, pp. 265–322.
- Ray B (1993) Sublethal injury, bacteriocins, food microbiology. ASM News 56: 285–29100. Schved F, Henis Y and Juven BJ (1994) Respone of spheroplasts and chelator–permeabilized cells of Gram–negative bacteria to the action of the bacteriocins pediocin SJ–1 and nisin. Int J Food Microbiol 21: 305–314.
- Cleveland J, Montville TJ, Nes IF, Chikindas ML (2001) Bacteriocins: safe, national antimicrobials for food preservation. Int J Food Microbiol 71: 1–20.
- Bromberg R, Moreno I, Zaganini CL, Delboni RR, de Oliveira J (2004) Isolation of bacteriocin–producing lactic acid bacteria from meat and meat products and its spectrum of inhibitory activity. Braz J Microbiol 35(1–2): 137-144.
- Parada JL, Caron CR, Bianchi A, Medeiros P, Soccol CR (2007) Bacteriocins from lactic acid bacteria: purification, properties and use as biopreservatives. Braz Arch Biol Tech 50(3): 521–542.
- Gardner HL (1983) Pathogenicity of Gardnerella vaginalis (Haemophilus vaginalis). Scand J Infect Dis Supply 40: 37–40.
- Romero R, Hagay Z, Nores J, Mazor M (1992) Eradication of urea plasma ureatyticum from the amniotic fluid with transplacental antibiotic treatment. Am J Obstet Gynecol 166: 618–620.
- Hill GB (1993) The microbiology of bacterial vaginosis. Am J Obstet Gynecol 169(22): 450–454.
- Mc Gregor JA, French JI, Parker R, Draper D, Patterson E, et al. (1995) Prevention of premature birth by screening and treatment for common genital tract infections; results of a prospective controlled evaluation. Am J Obstet Gynecol 173: 157–167.
- Hay PE, Morgan DJ, Ison CA, Bhide SA, Romney M, et al. (1994) A longitudinal study of bacterial vaginosis during pregnancy. Int J Obstet Gynaecol 101(12): 1048–1053.
- Holst E, Goffeng AR and Andersch B (1994) Bacterial vaginosis and vaginal microorganisms in idiopathic premature labor and association with pregnancy outcome. J Clin Microbiol 32: 176–186.
- Krohn MA, Hillier SL, Nugent RP, et al. (1995) The genital flora of woman with intra–amniotic infection. Vaginal infection and prematurity study group. J Infect Dis 71: 1475–1480.
- Ness RB, Hillier SL, Kip KE, Soper DE, Stamm CA, et al. (2004) Bacterial vaginosis and risk of pelvic inflammatory disease. Obstet Gynecol 104(4): 761–769.
- Lam MH, Birch DF and Fairley KF (1988) Prevalence of Gardnerella vaginalis in the urinary tract. J Clin Microbiol 26(6): 1130–1133.
- Martin HL, Richardson BA, Nyange PM, Lavreys L, Hillier SL, et al. (1999) Vaginal Lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J Infect Dis 180: 1863–1868.
- Cherpes TL, Meyne LA, Krohn MA, Hiller SL (2003) Risk factors for infection with herpes simplex virus type 2: role of smoking, douching, uncircumcised males, and vaginal flora. Sex Transm Dis 30: 405–410.
- Goldstein EJC, Citron DM, Cherubin CE, Hillier SL (1993) Comparative susceptibility of anaerobic bacteria to meropenem, imipenem, piperacillin, cefoxitin, ampicillin/ sulbactam, clindamycin and metronidazole. J Antimicrob Chemother 31: 363–372.
- Hay P (2000) Recurrent bacterial vaginosis. Curr Infect Dis Rep 2(6): 506–512.
- Graham DY, Adam E, Reddy GT, Agarwal JP, Agarwal R et al. (1991) Seroepidemiology of Helicobacter pylori infection in India comparison of developing and developed countries. Dig Dis Sci 36: 1084–1088.
- Smoot DT (1997) How does Helicobacter pylori cause mucosal damage? Direct mechanisms. Gastroenterol 113: 31–34.
- Perna F, Zullo A, Ricci C et al. (2007) Levofloxacin–based triple therapy for Helicobacter pylori re–treatment: role of bacterial resistance. Dig Liver Dis 39: 1001–1005.
- Hsu PI, Wu DC, Chen A, Peng NJ, Tseng HH, Tsay FW, et al. (2008) Quadruple rescue therapy for Helicobacter pylori infection after two treatment failures. Europ J Clin Investig 38(6): 404–409.
- Björkholm B, Sjölund M, Falk PG, Berg OG, Engstrand L, et al. (2001) Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc Natl Acad Sci USA 98: 14607–14612.
- Kim MJ and Kao C (2001) Factors regulating template switch in vitro by viral RNA–dependent RNA polymerases: implications for RNA–RNA recombination. Proc Natl Acad Sci USA 98: 4972–4977.
- Megraud F (2004) H. pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut 53: 1374–1384.
- Fallahi GH and Maleknejad S (2007) Helicobacter pylori culture and antimicrobial resistance in Iran. Indian J Pediatr 74: 127– 130.
- Bago J, Majstorovi? K, Beloši?–Halle Z, Ku?išec N, Bakula V, Tomi? M, et al. (2010) Antimicrobial resistance of H. pylori to the outcome of 10–days vs. 7–days moxifloxacin based therapy for the eradication: a randomized controlled trial. Ann Clin Microbiol Antimicrob 9: 13.
- Zárate G, Santos V, Nader–Macias ME (2007) Protective effect of vaginal Lactobacillus paracasei CRL1289 against urogenital infection produced by Staphylococcus aureus in a mouse animal model. Infect Dis Obstet Gynecol 2007: 48358–48363.
- Skarin A and Sylwan J (1986) Vaginal lactobacilli inhibit growth of Gardnerella vaginalis, Mobiluncus and other bacterial species cultured from vaginal content of women with bacterial vaginosis. Acta Pathol Microbiol Immunol Scand Sect B 94: 399–403.
- Kim TS, Hur JW, Yu MA, Cheigh CI, Kim KN, et al. (2003) Antagonism of Helicobacter pylori by bacteriocins of lactic acid bacteria. Department of Biotechnology and Bioproducts Research Center, Yonsei University, Seoul 120–749, Korea.
- Simova ED, Beshkova DB and Dimitrov ZHP (2009) Characterization and antimicrobial spectrum of bacteriocins produced by lactic acid bacteria isolated from traditional Bulgarian diary products. J Appl Microbiol 106: 692–701.
- Metchinkoff E (1908) The Prolongation of Life. Putmans Sons, New York, pp. 151–183.
- Kollath W (1953) Ernährungund Zahnsystem. Deutsche Zahnärztliche Zeitschrift 8: 7–16.
- Fuller R (1992) Probiotics. The scientific basis. London: Chapman & Hall.
- Anuradha S and Rajeshwar K (2005) Probiotics in health and disease. JIACM 6(1): 67–72.
- Liong MT (2008) Roles of probiotics and prebiotics in colon cancer prevention: postulated mechanisms and in–vivo evidence. Int J Mol Sci 9(5): 854–863.
- Choi NW, Shettigara PT, Abu–Zeid HAH, Nelson NA (2006) Herpes virus infection and cervical anaplasia–a seroepidemiological study. Intern J Cancer 19(2): 167–171.
- Nagy E, Petterson M and Mardh PA (1991) Antibiosis between bacteria isolated from the vagina of women with and without signs of bacterial vaginosis. APMIS 99: 739–744.
- Redondo–Lopez V, Cook RL and Sobel JD (1990) Emerging role of lactobacilli in the control and maintenance of the vaginal bacterial microflora. Rev Infect Dis 12: 856–872.
- Wilson M (2005) The reproductive system and its indigenous microbiota. In: Microbial inhabitants of humans: their ecology and role in health and disease. Cambridge, UK, Cambridge University Press, pp. 6–250.
- Zhou X, Bent ST, Schneider MG, Davis CC, Islam MR, Forney LJ ( 2004) Characterization of vaginal microbial communities in adult healthy women using cultivation–independent methods. Microbiol 150: 2565–2573.
- Ocaña VS, de Ruiz Holgado AAP, Nader–Macías ME (1999) Characterization of a bacteriocin–like substance produced by a vaginal Lactobacillus salivarius strain. Appl Environ Microbiol 65(12): 5631–5635.
- Aroutcheva AA, Simoes JA and Faro S (2001) Antimicrobial protein produced by vaginal Lactobacillus acidophilus that inhibits Gardnerella vaginalis. Infect Dis Obstet Gynecol 9: 33–39.
- Boris S and Barbès C (2000) Role played by Lactobacillus in controlling the population of vaginal pathogens. Microbes Infect 2: 543–546.
- Turovskiy Y, Ludescher RD, Aroutcheva AA, Faro S, Chikindas ML (2009) Lactocin 160, a Bacteriocin produced by vaginal Lactobacillus rhamnosus, targets cytoplasmic membranes of the vaginal pathogen, Gardnerella vaginalis. Probiotics Antimicrob Protein 1(1): 67–74.
- Kabir AM, Aiba Y, Takagi A, Kamiya S, Miwa T, et al. (1997) Prevention of Helicobacter pylori infection by lactobacilli in a gnotobiotic murine model. Gut 41: 49–55.
- Michetti P, Dorta G, Wiesel PH, Brassart D, Verdu E, et al. (1999) Effect of whey–based culture supernatant of Lactobacillus acidophilus (johnsanii) La1 on Helicobacter pylori infection in humans. Dig 60: 203–209.
- Armuzzi A, Cremonini F, Bartolozzi F, Canducci F, Candelli M, et al. (2001) The effect of oral administration of Lactobacillus GG on antibiotic–associated gastrointestinal side–effects during Helicobacter pylori eradication therapy. Aliment Pharmacol Ther 15: 163–169.
- Canducci F, Cremonini F, Armuzzi A, et al. (2001) Probiotics and Helicobacter pylori eradication. Dig Liver Dis 34: 81–83.
- Lorca GL, Wadstrom T, Valdez GF, Ljungh A (2001) Lactobacillus acidophilus autolysis inhibitHelicobacter pylori in vivo. Curr Microbiol 42: 39–44.
- Pinchuk IV, Bressollier P, Verneuil B, Fenet B, Sorokulova IB, et al. (2001) In vitro anti–Helicobacter pylori activity of the probioticstrain Bacillus subtilis 3 is due to secretion of antibiotics. Antimicrob Agents Chemother 45: 3156–3161.
- Sakamoto I, Igarashi M, Kimura K, Takagi A, Miwa T, et al. (2001) Suppressive effect of Lactobacillus gasseri OLL 2716 (LG21) on Helicobacter pylori infection in humans. J Antimicrob Chemother 47: 709–710.
- Nam H, Ha M, Bae O, Lee Y (2002) Effect of Weissella confusa strain PL9001 on the adherence and growth of Helicobacter pylori. Appl Environ Microbiol 68: 4642–4645.
- Sheu BS, Wu JJ, Lo CY, et al. (2002) Impact of supplement with Lactobacillus and Bifidobacterium containing yogurt on triple therapy for Helicobacter pylori eradication. Aliment Pharmacol Ther 16: 1669–1675.
- Cats A, Kuipers EJ, Bosschaert MA, Pot RG, Vandenbroucke– Grauls CM, et al. (2003) Effect of frequent consumption of a Lactobacillus casei–containing milk drink in Helicobacter pylori– colonized subjects. Aliment Pharmacol Ther 17: 429–435.
- Linsalata M, Russo F, Berloco P, Caruso ML, Matteo GD, et al. (2004) The influence of Lactobacillus brevis on ornithine decarboxylase activity and polyamine profiles in Helicobacter pylori–infected gastric mucosa. Helicobacter 9: 165–172.
- Nista EC, Candelli M, Cremonini F, et al. (2004) Bacillus clausii therapy to reduce side–effects of anti–Helicobacter pylori treatment: randomized, double–blind, placebo controlled trial. Aliment Pharmacol Ther 20: 1181–1188.
- Sgouras D, Maragkoudakis P, Petraki K, Martinez–Gonzales B, Eriotrou E, et al. (2004) In vitro and in vivo inhibition of Helicobacter pylori by Lactobacillus casei strain shirota. Appl Environ Microbiol 70(1): 518–526.
- Tursi A, Brandimarte G, Giorgetti GM, Forti G, Modeo ME, et al. (2004) Low–dose balsalazide plus a high–potency preparation is more effective than balsalazide alone or mesalazine in the treatment of acute mild–tomoderate ulcerative colitis. Med Sci Monit 10(11): 126–131.
- Wang G and Maier RJ (2004) An NADPH quinone reductase of Helicobacter pylori plays an important role in oxidative stress resistance and host colonization. Infect Immun 72: 1391–1396.
- Johnson–Henry KC, Nadjafi M, Avitzur Y, Mitchell DJ, Nran BY, et al. (2005) Amelioration of the effects of Citribacter rodentium infection in mice by pretreatment with probiotics. J Inf Dis 191: 2106–2117.
- Pena JA, Rogers AB, Ge Z, Ng V, Li SY, et al. (2005) Probiotic Lactobacillus spp. diminishes Helicobacter hepaticus–induced inflammatory bowel disease in interleukin–10–deficient mice. Infect Immun 73: 912–920.
- Kieran AR, Daly P, Li Y, Hooton C, Paul W (2008) Strain–specific inhibition of Helicobacter pylori by Lactobacillus salivarius and other lactobacilli. J Antimicrob Agents Chemother 61(4): 831–834.
- Ray SK, Johnson MC and Ray B (1989) Bacteriocin plasmids of Pediococcus acidilactici. J Indust Microbiol 4: 163–179.
- Ray SK, Kim WJ, Johnson MC, Ray B (1989) Conjugal transfer of a plasmid encoding bacteriocin production and immunity in Pediococcus acidilactici H. J Appl Bacteriol 66: 393–399.
- Kim WJ, Ha DM and Ray B (1991) Characteristics of bacteriocin and mucin production phenotypes in Lactobacillus plantarum 27. J Microbiol Biotechnol 1: 96–101.
- Ray B, Motlagh AM, Johnson MC, Bozoglu (1992) Mapping of pSMB74, a plasmid–encoding bacteriocin pediocin AcH production (Pap+) by Pediococcus acidilacticl H. Lett Appl Microbiol 15: 35–37.
- Nettles CG and Barefoot SF (1993) Biochemical and genetic characterization of bacteriocin of food associated lactic acid bacteria. J Food Prot 56: 338–356.
- Kantor A, Montville TJ, Mett A, Shapira R (1997) Molecular characterization of the replicon of the Pediococcus pentosaceus 43200 pediocin A plasmid pMD136. FEMS Microbiol Lett 151: 237–244.
- Bukhtiyarova M, Yang R and Ray B (1994) Analysis of pediocin AcH gene cluster from plasmid pSMB74 and its expression in a pediocin–negative Pediococcus acidilactici strain. Appl Environ Microbiol 60: 3405–3408.
- Motlagh A, Bukhtiyarova M and Ray B (1994) Complete nucleotide sequences of pSMB74, a plasmid encoding production of pediocin AcH in Pediococcus acidilactici. Lett Appl Microbiol 18: 305–312.
- Miller KW, Schamber R, Chen Y, Ray B (1998) Production of active chimeric pediocin AcH in Escherichia coli in the absence of processing and secretion genes from the Pediococcus Pap operon. Appl Environ Microbiol 64: 14–20.
- Henderson JT, Chopko AL, van Wassenaar PD (1992) Purification and primary structure of pediocin PA–1 produced by Pediococcus acidilactici PAC1.0. Arch Biochem Biophys 295: 5–12.
- Miller KW, Schamber R, Osmanagaoglu O, Ray B (1998) Isolation and characterization of pediocin AcH chimeric protein mutants with altered bactericidal activity. Appl Environ Microbiol 64: 1997–2005.
- Tagg JR, Dajani AS and Wannamaker LW (1976) Bacteriocins of gram–positive bacteria. Bacteriol Rev 40: 722–756.
- Daeschel MA (1989) Antimicrobial substances from lactic acid bacteria for use as food biopreservatives. Food Technol 43: 164–167.
- Stiles ME and Hastings JW (1991) Bacteriocin production by lactic acid bacteria: potential for use in meat preservation. Food Sci Technol 2: 235–263.
- Klaenhammer TR (1993) Genetics of bacteriocin production by lactic acid bacteria. FEMS Microbiol Rev 12: 39–86.
- Papagianni M and Anastasiadou S (2009) Pediocins: The bacteriocins of Pediococci. Sources, production, properties and applications. Microb Cell Fact 8: 3. doi-10.1186/1475-2859-8-3.
- Coderre PE and Somkuti GA (1999) Cloning and expression of the pediocin operon in Streptococcus thermophilus and other lactic fermentation bacteria. Curr Microbiol 39(5): 295–301.
- Moon Gi–S, Pyun Yu–R and Wang JK (2006) Expression and purification of a fusion–typed pediocin PA–1 in Escherichia coli and recovery of biologically active pediocin PA–1. Int J Food Microbiol 108: 136–140.
- Beaulieu L, Tolkatchev D, Jetté JF, Groleau D, Subirade M (2007) Production of active pediocin PA–1 in Escherichia coli using a thioredoxin gene fusion expression approach: cloning, expression, purification, and characterization. Can J Microbiol 53(11): 1246–1258.
- Halami PM and Chandrashekar A (2007) Heterologous expression, purification and refolding of an anti–listerial peptide produced by Pediococcus acidilactici K7. Electron J Biotechnol 10(4) doi:10.2225.
- Chikindas ML, Venema K, Ledeboer AM, Venema K, Ledeboer AM, et al. (1995) Expression of lactococcin A and pediocin PA–1 in heterologous hosts. Lett Appl Microbiol 21(3): 183–189.
- Horn N, Martinez MI, Martinez JM, Hernandez PE, Gasson MJ, et al. (1998) Production of pediocin PA–1 by Lactococcus lactisusing the lactococcin A secretory apparatus. Appl Environ Microbiol 64: 818–823.
- Hoartinez MI, Martinez JM, Hernandez PE, Gasson MJ, Rodriguez JM, et al. (1999) Enhanced production of pediocin PA–1 and co–production of nisin and pediocin PA–1 by Lactococcus lactis. Appl Environ Microbiol 65(10): 4443–4450.
- Martinez JM, Kok J, Sanders JW, Hernandezi PE (2000) Heterologous coproduction of enterocin A and pediocin PA–1 by lactococcus lactis: detection by specific peptide–directed antibodies. Appl Environ Microbiol 66(8): 3543–3549.
- Somkuti GA and Steinberg DH (2003) Agarose/agar assay system for the screening of bacteriocin–producing lactic fermentation bacteria. Biotech Lett 24: 303–308.
- Horn N, Ferna´ndez A, Dodd HM, Gasson MJ, Rodríguez JM (2004) Nisin–controlled production of pediocin PA–1 and colicin V in nisin and non–nisin–producing Lactococcus lactis strains. Appl Environ Microbiol 70(8): 5030–5032.
- Halami S and Prakash M (2008) Cloning of pediocin PA–1 and its immunity genes from Pediococcus acidilactici K7 using pAMJ shuttle vector into Lactococcus lactis MG1363. Indian J Biotechnol 7(4): 550–553.
- Arqués JL, Rodríguez JM, Gasson MJ, Horn N (2008) Immunity gene pedB enhances production of pediocin PA–1 in naturally resistant Lactococcus lactis strains. J Dairy Sci 91(7): 2591–2594.
- Johnsen L, Fimland G and Nissen–Meyer J (2005) The C– terminal domain of pediocin–like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the c–terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J Biol Chem 280(10): 9243–9250.
- Moon G–S, Pyun Y–R, Park MS, Geun Eog JI, Wang KIM (2005) Secretion of recombinant pediocin PA–1 by Bifidobacterium longum, using the signal sequence for Bifidobacterial α– amylase. Appl Environ Microbiol 71(9): 5630–5632.
- Schoeman H, Vivier MA, duToit M, Dicks LMT, Pretorius IS (1999) The development of bactericidal yeast strains by expressing the Pediococcus acidilactici pediocin gene (pedA) in Saccharomyces cesevisiae. Yeast 15(8): 647–656.193.
- Beaulieu L, Groleau D, Miguez CB, Jette JF, Aomari H, et al. (1999) Production of pediocin PA–1 in the methylotrophic yeast Pichia pastoris reveals unexpected inhibition of its biological activity due to the presence of collagen–like material. Protein Expr Purifi 43(2): 111–125.
- Buyong N, Kok J and Luchansky JB (1998) Use of a genetically enhanced, pediocin–producing starter culture, Lactococcus lactis subsp. lactis MM217, to control Listeria monocytogenes in Cheddar Cheese. Appl Environ Microbiol 64: 4842–4845.
- Tominaga T and Hatakeyama Y (2007) Development of innovative pediocin PA–1 by DNA shuffling among class IIa bacteriocins. Appl Environ Microbiol 73(16): 5292–5299.
- Grodberg J and Dunn JJ (1988) ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol 170: 1245–1253.
- Prinz WA, Aslund F, Holmgren A, Beckwith J (1997) The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in Escherichia coli cytoplasm. J Biol Chem 272: 15661–15667.
- Bessette PH, Aslund F, Beckwith J, Georgiou G (1999) Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci USA 96: 13703– 13708.
- Novy R, Drott D, Yaeger K, Mierendorf R (2001) Overcoming the codon bias of E. colifor enhanced protein expression. Innovations Newsletter of Novagen, Number 12.
- Makrides SC (1996) Strategies for achieving high–level expression of genes in Escherichia coli. Microbiol Mol Biol Rev 60(3): 512–538.
- Johnson MC, Hanlin MB and Ray B (1992) low pH and lactate are necessary for conversion of prepediocin to active pediocin AcH in Pediococcus acidilactici H, abstr. 0–81. In: Abstracts of the 92nd general meeting of the American Society for Microbiology, ASM, Washington.
- Motlagh AM, Holla S, Johnson MC, Ray B, Field RA (1992) Inhibition of Listeria spp. in sterile food systems by pediocin AcH, a bacteriocin from Pediococcus acidilactici H. J Food Prot 55: 337–343.
- Venema K, Kok J, Marugg JD, Toonen MY, Ledeboer AM, et al. (1995) Functional analysis of the pediocin operon of Pediococcus acidilactici PAC1.0 :PedB is the immunity protein and PedD is the precursor processing enzyme. Mol Microbiol 17: 515–522.
- Eijsink VGH, Skeie M, Middelhoven PH, Brurberg MB, Nes IF (1998) Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl Environ Microbiol 64: 3275–3281.
- Kane JF and Hartley DL (1991) Properties of recombinant protein–containing inclusion bodies in Escherichia coli. Bioproc Technol 12: 121–145.
- Mukhopadhyay A (1997) Inclusion bodies and purification of proteins in biologically active forms. Adv Biochem Eng Biotechnol 56: 61–109.











