Keywords
Coronavirus; Transmission; Diagnoses; Clinical features; Treatment; Vaccine; Spike; ACE
Abbreviations
SARS-CoV-2: New Beta-coronavirus discovered in Wuhan, Hubei province, China in December 2019; 2019-nCoV (2019 Novel Coronavirus): Refers the same virus as above and was initially used by WHO; HCoV-19: Alternate name used for the same virus as above; COVID-19 (Coronavirus Disease 2019): The disease caused by SARS-CoV-2 Infection; SARS: Severe Acute Respiratory Syndrome; SARS-CoV: Refers to the virus that causes SARS; MERS: Middle East Respiratory Syndrome; MERS-CoV: Refers to the virus that causes MERS; ACE: Angiotensin Converting Enzyme; RT-PCR: Reverse Transcriptase-Polymerase Chain Reaction; HRCT: High Resolution Computed Tomography; CXR: Chest X-Ray
Introduction
The usual, bustling streets of Wuhan during the 2020 Chinese New Year were now eerily quiet and empty, some might even say “ghostly”. The COVID-19 (Coronavirus Disease 2019) outbreak hit the people of Wuhan and the people of China in what was to be their most euphoric time-of-year- it also turned out to be one of their most vulnerable. Ninety days in, we perform a systematic review on information we have obtained on COVID-19 in this limited, yet crucial period [1-25].
As of March 18th, 2020, what had started as a regional outbreak in Wuhan and middle China in December 2019 has now spread to over 165 countries and regions including the U.S., infecting more than 200,000 people and resulting in more than 8000 deaths [26-37]. The WHO has officially declared the COVID-19 outbreak to be a global pandemic [38-53]. A possible source of the outbreak has been traced to a busy Huanan seafood market in Wuhan [54]. Allegedly, the seafood market was a trading place for various kinds of wild game, some of which have been traditional delicacies of the region for hundreds of years. According to folk beliefs, the meats of the wild game have natural healing powers and are thus often used in “dietary therapy” and traditional Chinese medicine. One such belief was that the consumption of pangolin could help improve circulation. Pangolins are also on the list of possible intermediate hosts for the SARS-CoV-2 [29]. Current epidemiological evidence suggest that bats are the most likely primary hosts for SARS-CoV-2 and that the development of human-to-human transmission allowed for the subsequent rapid viral spread [48,55-65]. Bats are often traded in China as an ingredient for traditional medicines, but they have often been the “notorious” primary hosts for various deadly viruses in history [66]. This “viral richness” of bats can be attributable to their elevated body temperature (compared to other mammals) as well as their unique and ultra-effective immune system. This results in a type of viral selection in bats where only the most virulent survive to transmit to other organisms or species [39].
The preliminary epidemiology data obtained in the first ninety days of the outbreak can provide a glimpse into the various characteristics of SARS-CoV-2. It was originally believed that the virus had limited human-to-human transmission capacities, but it has recently been shown that humans seem to be susceptible to the virus through droplet transmission or direct and indirect contact with contaminated surfaces. The incubation period has typically been observed to range from 2-14 days, with certain cases ranging up to 27 days; the basic reproductive number has been estimated to be around 2-3, although the real number may turn out to be much higher; And the mortality rate has come to around 2.38%, with men and immunocompromised patients having poorer prognosis [54]. For patients that succumbed to the virus, the median number of days from initial symptoms to death was 14 days, with a shorter period for patients above the age of 70 [64]. More information on the transmission mechanism of SARS-CoV-2 can be found in the corresponding section of this review. Fever and coughs are the most common presenting symptoms of patients infected with SARS-CoV-2 [64,67,68]. The most common diagnosis tool has been quantitative reverse transcriptase PCR (qRT-PCR) but medical professionals have been pushing to establish diagnostic criteria based on high definition chest CT, in order to circumvent the limited capacity of the PCR kits [69]. There are currently no specific treatments officially FDAapproved for COVID-19 other than general supportive therapy. However, multiple anti-viral drugs are going through clinical trials and vaccine development is underway [57]. The diagnosis process, the treatment, and the prevention of COVID-19 will be discussed in individual sections later in this review.
Following the WHO announcement on March 11th that upgraded the COVID-19 outbreak to a global pandemic, coordinated efforts into the research and treatment of this emerging virus have been gaining more attention and traction [38,60]. Although mainland China has taken on the vast majority of cases so far, other nations are also tightening up their public health defenses and starting to contribute to the process of understanding the virus and ultimately, finding a cure. For example, Japan, one of the most severely affected countries other than China, is also the host nation for the upcoming 2020 Summer Olympics [21]. Thus, Japan is under great pressure to keep the epidemic in check ahead of the world’s greatest support event, where heavy international traffic may exacerbate the spread of the virus [51].
This review seeks to capture the essence of the knowledge and information that we have learned about COVID-19 in these initial ninety days and hopefully inspire more work to be done in areas yet uncharted.
Transmission
Currently, SARS-CoV-2 is hypothesized to be transmitted through large water droplets [49]. This could explain the why the wet fish market in Wuhan was the epicenter for COVID-19 [49]. Aerosol transmission is also a possibility [49,70-82]. However, some coronaviruses, including but not limited to MERS-CoV, SARS-CoV, are transmitted through the oral-fecal route via intestinal infection and whether 2019-nCoV follows this transmission will need to be further tested [83]. However, Zhang et al. [80] was able to obtain more anal swab positives than oral swab positives during a later stage of the infection [83]. The virus was also found in blood [83]. This suggests patients can transmit this pathogen through three mechanisms: respiratory, fecal-oral, or body fluids [83]. In persons infected with SARS-CoV, angiotensin-converting enzyme 2 (ACE2) was mutated and could exhibit decreased expression of antimicrobial peptides [23]. This mutation Since ACE2 is found in gut microbial and mutations and ACE2 is mutated in SARS CoV patients, fecal-oral transmission is possible for general coronaviruses [23]. Therefore, there is a speculation that SARSCoV- 1 and SARS-CoV-2 have similar transmissions, since they are both coronaviruses. Gao et al. [22] suggests further studies to be done on how ACE2 affects transmission of coronaviruses (see section VII, Molecular Characteristics for details) [23].
Additionally, both SARS-CoV-1 and MERS-CoV were transmitted through the oral-fecal route via intestinal infection and whether or not the SARS-CoV-2 also follows this transmission will need to be further tested [83]. COVID-19 patients show gastrointestinal symptoms which further support the oral-fecal transmission hypothesis [79]. In areas with poor sanitation, precautions must be observed when handling stools of patients infected with SARSCoV- 2. In 2002, SARS-CoV-1 RNA found in sewage water at 4 °C remained infectious for 14 days in Beijing hospitals treating patients with SARS [79]. Nations with high urban populations and poor sanitation measures are thus potentially more susceptible to transmission of SARS-CoV-2. Further research will need to be done regarding the possibility of the oral-fecal route including environmental studies [79].
It was previously suggested that the transmission of SARSCoV- 2 could not be from an asymptomatic carrier [4]. However, this is incorrect as Bai et al. [4] were able to determine that 5 patients had contact with an asymptomatic patient infected with COVID-19. The incubation period for this asymptomatic patient was 19 days and her first RT-PCR result was negative. Further proof of study will be needed to accurately determine the extent to which asymptomatic carriers could acquire and transmit SARS-CoV-2.
Healthcare providers and clinicians must be prepared to provide treatments, especially providing specific organ treatments to COVID-19 patients [49]. Extracorporeal therapies can be helpful for treatment of the lungs, heart, kidneys, and liver. Extracorporeal membrane oxygenation (ECMO) can be used to treat acute hypoxemic respiratory failure [49]. It is also important to consider the transmission data when predicting outcomes of SARS-CoV-2. Table 1 summarizes each studies’ transmission data.
Table 1 Summary of each studies’ transmission data.
| |
Studies |
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
| Mean incubation period (days) |
6.4 |
5.5 |
5.2 |
|
6.0* |
5.0* |
5.1** |
| Doubling time (days) |
|
7.31 |
7.4 |
|
6.4 |
|
|
| Serial Interval (days) |
|
|
7.5 |
|
8.4* |
7.6* |
|
| Basic Reproductive Number, Ro (persons) |
|
|
2.2 |
3.28 |
2.68 |
2.24-3.58 |
2.8-3.3 |
Note. All studies presented a 95% CI except when noted:
*Assumption made consistent with SARS-CoV and MERS-CoV (95% CI).
**Inferred infection during free propagation of 2019-nCoV and used recent reports (95% CI).
The age range for infected patients as reported by Backer et al. [3] is 2 - 72 years old and Li et al. [30] is 15 - 89 years old. Li et al. [30] reported that 56% of the infected subjects in their study were male (n=425) and 55% of the reported cases studied were before January 1st, 2020. Reports have shown that it may affect males more the females, but there has been no definite proof found. More male patients have reported exposure to the Huanan Seafood Wholesale Market. (Li) Also, Channappanavar et al. [8] found that male mice were more susceptible to SARSCoV infection compared with age-matched females. Male mice had higher virus titers, enhanced vascular leakage, and alveolar edema. There seems to be a protective effect for estrogen receptor signaling in mice infected with SARS-CoV, making females less susceptible to infection. To our knowledge, there is a lack of research on the susceptibility of different racial ethnicities to being infected by SARS-CoV-2.
The mean incubation period refers to the time from exposure to onset of symptoms. The incubation period between the 7 studies had a range of 5.0 - 6.4 days [3,18,30,34,71,84-86]. Excluding the 4 studies that didn’t present data or presented assumed data from previous literature, the mean incubation period was 6.4 days, 5.5 days, and 5.2 days, for Backer et al. [3] Du et al. [17] and Li et al. [28] respectively.The CDC reports the incubation period as 2 - 14 days [26]. Backer et al. [3] had three parametric models to predict the incubation period distribution (weibull, gamma, and lognormal). The weibull model had the best fit while the gamma and lognormal had a higher range (2.4 - 12.5 and 2.4 - 15.5, respectively) [3]. The study was limited by selective sampling (more males and younger population) and only on confirmed cases. Li et al. [30] reports that in addition to the 5.2 days of mean incubation period, their distribution provides strong evidence to support a 14-day medical observation only for confirmed cases (two positive results from RT-PCR). This quarantine period is crucial for exposed patients so as to prevent further transmission of the viral infection. Although the Li et al. [30] studies were limited by a lack of diagnostic reagents, they state that urgent next steps need to be identified immediately to effectively reduce the transmission of SARS-CoV-2. All data presented had a 95% CI.
The doubling time is the time it takes to double the number of cases in a certain population. The doubling time was found to be 7.31 days, 7.4 days, and 6.4 days for Du et al. [18], Li et al. [30], and Wu et al. [71], respectively. Wu et al. [71] reported limitations to their study such as uncertainty regarding the lag time between infection and case detection, which could hinder accurate data collection. Wu et al. [71] was able to forecast the global spread of SARS-CoV-2from Wuhan. The validity of their study was supported by a consensus of the same forecast methodology of other studies. All data presented had a 95% CI.
The serial interval refers to the duration between two successive patient cases in a transmission chain. Only 1 transmission study out of the 7 studies we reviewed calculated the serial interval value (2 other studies used assumed values). Li et al. [28] found that the serial interval was 7.5 days [30]. This was very similar to the doubling time presented, as expected. This means that the symptoms appear soon after a person is infected with SARSCoV- 2. All data presented had a 95% CI.
One of the most important statistical values that were found in 5 of the 7 studies was the basic reproductive number (Ro). Ro is the expected number of cases that are directly generated by one patient with the viral infection. Li et al. [29] found the lowest average basic reproductive number at 2.2 persons while Zhao et al. [85] found the highest average basic reproductive number at 3.58 persons [30]. As shown by the large value discrepancy, there has yet to be a solid consensus on this topic. WHO reports the basic reproductive number for COVID-19 to be between 1.4–2.5 persons, although the real number may be much higher [34]. The 5 studies we reviewed presented overall higher Ro values compared to WHO’s estimate. However, there are some concerns with the Ro value found in the studies. Due to short onset time, the current estimates may be biased. Zhoa et al. [86] found that Ro was the most sensitive to the generation time (Tg). Tg is the average length of time from the release of the virion until it infects another cell and causes the release of another virion. The smaller the value of Tg, the higher the To value. A sensitivity analysis was also conducted to determine the best-case and worst-case scenarios in calculating the Ro. The best-case scenario was 2.3 persons while the worst-case scenario was 5.2 persons [86].This sensitivity analysis adjusted for a range of Tg between 8-12 days.
All data presented had a 95% CI.
Very few studies presented outcome models for COVID-19 on young children because most patients infected so far were adults and the elderly. However, vertical transmission (i.e. transplacental, birth canal, human breast milk) could still be possible and has not been ruled out. Continued research will need to be done to ascertain the transmission of SARS-CoV-2 to pediatric patients. Wei et al. [67] reported that nine infants (age under 1 years old) were identified to be infected with SARS-CoV-2 between December 8th, 2019 and February 6th, 2020. They all had at least 1 infected family member and 7 out of the 9 infants were female. There were limitations to this study, due to the small sample size and the lack of inclusion of the asymptomatic patients. A suggestion that Wei et al. [67] presented when treating infants with COVID-19 is to have them wear specific protective masks that fit their facial size.
The results presented are comparable to previous viral infections and it is important to continue surveillance efforts to prevent global pandemic [83]. There are a few future questions that researchers must ask regarding transmissibility of this agent. Lipsitch et al. [33] suggested these questions to be asked: What is the full spectrum of COVID-19 severity? Who are the specific infector’s profiles? What are the risk factors for severe illness or death? The main recommendation for viral testing is to reserve the testing capacity to support public health efforts to characterize the severity and trajectory of the disease. Additional questions include when and how the virus emerged and who is patient zero? Is the Huanan market the true ground zero? The main recommendation for viral testing is to reserve the testing capacity to support public health efforts to characterize the severity and trajectory of the disease.
Diagnosis and Clinical Features
Before the details of the diagnosis and clinical features of COVID-19 is discussed, it is important to take note of the patient population. In two separate studies performed on 237 total patients by Jinyintan hospital and Zhongnan Hospital in Wuhan, the median patient age was around 55-56, with more male than female patients [10]. Researchers from Zhongnan hospital also noted that the median time from a patient’s initial symptoms to his/her hospital admission was 7 days and patients who needed to be admitted to the ICU were on average 11 years older than the general patient population. Among the recovered patients, the median length-of-hospital-stay was 10 days [61].
A great challenge in differentiating a COVID-19 infection from other pneumonias on physical exam is perhaps due to the former’s lack of specific signs and symptoms. Covid-19 patients often present with similar signs and symptoms to that of the common flu. Among the patient cohort studied by Tongji hospital in China, the most prevalent presenting symptom was fever. Other common non-specific signs and symptoms included cough, shortness of breath and muscle aches/fatigue [10]. In terms of lab test findings, Wang et al. [61] found that more than half of the 138 Wuhan patients recorded in the study presented with lymphopenia and prolonged PT (70.3% and 58% respectively). Elevated lactate dehydrogenase was also seen in 39.9% of patients, perhaps indicating an increase in cellular anaerobic respiration in response to infection [61]. Increased levels of hypersensitive C reactive protein (hs-CRP) was seen in twenty-seven out of the twenty-nine patients studied at Tongji hospital, indicative of an inflammatory process. Fifteen patients in the same study cohort also had decreased serum albumin levels, but interestingly, other lab values that would indicate a possible hepatic or renal disease process (ALT/AST/Total bilirubin/Serum Creatinine) were not elevated. In addition to the commonly measured lab values, the Interleukin-2R and Interleukin-6 expression levels in the serum of COVID-19 patients have been shown to correlate with the severity of disease [10]. This may be a helpful indicator for physicians to look to for a quick, preliminary assessment of the progression of the disease. Viral load was studied by Zou et al. [87,88] who demonstrated that levels tended to be higher in the samples collected from patients’ noses rather than their throats. Crucially, they also found that there was no difference in viral load measured in symptomatic vs. asymptomatic patients, likely suggesting a consistent transmission potential of COVID-19 across patient populations. See Table 2 for a summary on common signs and symptoms, as well as lab test findings for COVID-19 patients.
Table 2 On Common Signs & Symptoms as well as Lab Test Findings for COVID-19 Patients.
| Common Signs & Symptoms |
Common Lab Test Findings |
| Fever* |
Lymphopenia* |
| Cough (Typically Dry)* |
Prolonged Prothrombin Time (PT)* |
| Fatigue* |
Elevated Lactate Dehydrogenase* |
| Shortness of Breath (SOB) |
Elevated Hypersensitive C-Reactive Protein (hs-CRP) |
| Muscle Aches |
Decreased Serum Albumin |
| Confusion |
Elevated Serum Interleukin-2R and Interleukin-6 |
| Headache |
|
| Sore throat |
|
| Rhinorrhea |
|
*Indicates Signs/Symptoms/Lab Test findings that were present in>50% of studied patients (Compiled from [9,10,61]).
In addition to the discussion of typical presenting symptoms of COVID-19, it is also important to be aware that patients can be asymptomatic yet carry sufficient viral load to result in transmission. This was shown by Pan et al. [42] in a study performed on a family of three, all of whom were infected with the 2019-nCoV. It was only upon one of them showing symptoms that all three got tested via RT-PCR. The two other members remained asymptomatic and had normal lab values at admission. This may help explain how some “superspreaders” are able to transmit to multiple other people without realizing that they are infected.
Recent research has also shown key differences in viral load patterns of the 2019-nCoV compared to other members of its viral family. Kim et al. [25] demonstrated in that viral loads of the SARS-CoV-2 in patients peaked in the early phases of the disease process (3-5 days after initial symptom onset), a distinct pattern not seen with the SARS-CoV. In fact, the 2019-nCoV viral loads peaked in the lower respiratory tract before any associated symptoms presented. This may have significant implications when designing the optimal time course of antivirals for COVID-19 patients. Another key finding was that viral loads of the 2019- nCoV collected from the upper and lower respiratory tracts were quite similar, unlike what is seen with the MERS-CoV, with viral loads concentrating in the lower respiratory tract.
With the physical exam and lab test findings being less effective than physicians would have liked at differentiating COVID-19 from other viral infections, a commonly used diagnostic method is qRTPCR [14]. It has been established that SARS-CoV-2 can reliably be obtained from patient saliva, this allows for convenient collection of specimens for qRT-PCR [59]. Interestingly, in the development of the qRT-PCR probes for COVID-19, the RNA of the SARSCoV- 1 was used as a positive control. Researchers subsequently produced a RdRP_SARSr-P2 probe that specifically reacted with SARS-CoV-2, allowing for its differentiation from the SARS-CoV-1 [14]. The qRT-PCR is designed to detect genes of COVID-19 responsible for the RNA-dependent RNA polymerase as well as the Spike (S) protein on the viral surface [7]. Another design for qRT-PCR targets the Orf1b and N-region of the COVID-19 genome [16]. These designs were once again modeled after the qRT-PCR test for SARS-CoV-1, which had similar characteristics. Despite being such an effective diagnostic tool, the qRT-PCR comes not without its disadvantages, such as high rates of false-negatives [19]. Currently, the growing number of cases emerging in mainland China per day may temporarily overwhelm the nation’s capacity to perform such an expensive and sophisticated test. Thus, many physicians and researchers alike are pushing for the establishment of new diagnostic criteria of COVID-19 based on high resolution CT (HRCT) imaging results.
An imaging study of 63 patients at Tongji hospital showed that the most common signs on HRCT for COVID-19 patients were “patchy/ground glass opacities (GGO), patchy consolidations, fibrous stripes and irregular solid nodules”, ranked in terms of their respective prevalence [41]. It is important to note that GGOs are not specific radiological findings to COVID-19 patients, but how these findings progress and change throughout the course of disease may help clinicians identify the infection better. Subsequently, researchers at Tongji hospital were able to sequentialize the progression of these CT findings into 4 stages, with day one being the onset time of initial symptoms [41]. Stage I (0-4 days) consisted of the formation of GGOs in a majority of patients; Stage II (5-8 days) demonstrated a “crazy-paving pattern” with more lobar involvement; Stage III (9-13 days) marks the peak of lung involvement, typically at day 10, when GGOs and consolidations were at their maximum; Stage IV (>14 days) showed a gradual reduction in consolidations and the paving pattern [41]. Xu et al. [77] demonstrated the progression of the disease in bilateral lung lobes from day 2 to day 7.
Shi et al. [50] also presented similar findings on their patient’s CT images as he progressed through the course of disease, including a Chest X-Ray (CXR) obtained at admission. What is worth noting is that CXR findings are not as reliable as HRCT findings for COVID-19 patients. Normally, some infiltrates could be seen on CXR, but that’s not always the case. Zhang et al. [82] reported cases in Beijing where patients had normal CXRs but showed GGOs on HRCT.
Doctors at Second Xiangya Hospital in Changsha, China have documented five patients presenting with the symptoms of COVID-19 but had negative initial RT-PCR results. HRCTs of the patients were subsequently obtained, and all five patients showed similar GGO patches as described in cases above. Later, repeated swab tests confirmed their diagnoses of COVID-19 [75]. Furthermore, a study of 51 COVID-19 patients at Taizhou hospital demonstrated that HRCT achieved a diagnostic sensitivity of 98% compared to 71% of the qRT-PCR with p<0.001 [19]. Findings like these once again demonstrate the usefulness of HRCT in identifying COVID-19, even before an official diagnostic criterion is established.
Pathological slides have also been made obtained during biopsies of COVID-19 patients. In the lungs, the formation of hyaline membranes and diffuse pneumocyte damage was observed, indicative of Acute Respiratory Distress Syndrome (ARDS). Lymphocytic infiltrates and multinucleate syncytial cells were also seen, indicative of a viral pathologic process. In the liver, mild vascular steatosis was observed, similar to biopsy findings for the SARS-CoV and MERS-CoV, which could be induced by the virus or hepatotoxic drugs. Heart tissue biopsies were mostly normal in COVID-19 patients [78].
It is thus the hope of many medical professionals to quickly absorb and compile the radiological knowledge obtained from the growing base of patients into an effective and efficient diagnostic criterion that could be used universally. If such a radiological diagnostic protocol could be set up, it would greatly reduce the burden of many overworked genetics labs, where tens of thousands of patient RT-PCR specimens would have to be worked up. Diagnosis of COVID-19 by HRCT could be quicker, more accessible and most importantly, more sensitive. And in the field of infectious disease, time is invaluable.
Treatment
As of the March 2020, there are no FDA-approved drugs to treat SARS-CoV-2 or other coronaviruses. However, The CDC made a prescient promise, just one week prior to the COVID-19 outbreak, to preemptively prepare for the onset of a novel virus using new therapeutic and diagnostic technology [52]. Prior investments in health preparedness has prepared us well for new epidemics, but improvements must be made to accelerate discovery of treatments for future novel diseases. The current treatments and interventions for COVID-19 are compiled in Table 3 [12,17,22,28,32,40,56,62,72,80,84].
Table 3 Current treatments and interventions used for COVID-19 [12,17,22,28,32,40,56,62,72,80,84].
| Treatments |
Mechanism of Action |
| Immunoenhancers: Targets mainly SARS-CoV [13,80] |
| Interferons (IFN) |
IFN inhibits replication of animal and human coronaviruses Interferon-α2b is especially effective for pediatric patients |
| Intravenous gammaglobulin |
Immunomodulating drug for long-term use in all ages |
| Thymosin alpha-1 |
Thymocyte development and restores homeostasis of the immune system |
| Thymopentin |
Enhance antibody response |
| Levamisole |
Treatment of general infections |
| Cyclosporine A |
Inhibits replication of viruses that contain the nucleocapsid protein |
| Chinese medicine |
Enhance host immunity against infections of COVID-19 (i.e. glycyrrhizin, baicalin, ginseng stem-leaf saponins) |
| Coronavirus-specific treatments [22,62,80] |
| Chymotrypsin-like protease inhibitors, Papain-like protease (PLP) inhibitors |
Inhibit coronavirus encoded proteins that serve a function in replication and inhibition of host innate immune responses (i.e. Cinanserin, Flavonoids, PLP inhibitors, Diarylheptanoids, Spike protein ACE2 blockers, Human monoclonal Ab, Emodin, Promazin, Nicotianamine) |
| Chloroquine |
Block virus infection by increasing endosomal pH required for virus/cell fusion and interfering with glycosylation of cell receptors of SARS-CoV and possibly COVID-19. |
| Antiviral treatments [12,17,28,32,62,80] |
| Ribavirin |
Inhibition of the replication of SARS-associated coronavirus |
| Lopinavir/ritonavir |
Stimulates protease inhibitors in the treatment of viral infection Especially effective for pediatric patients |
| Remdesivir |
Remdesivir is an adenosine analogue, which incorporates into nascent viral RNA chains and results in pre-mature termination |
| Nelfinavir |
Selective inhibitor of viral proteases |
| Arbidol |
Russian-made small indole-derivative molecule that blocks viral fusion against influenza viruses |
| Nitric Oxide (NO) |
Inhibits the synthesis of viral protein and RNA |
| Other Compounds [40,56,80,84] |
| Alpha-lipoic acid |
Antioxidant |
| Estradiol and phytoestrogen |
Reduction of viral replication in primary human nasal epithelial cells derived from only female donors |
| Mucroporin-M1 |
Broad-spectrum virucidal activity against viruses |
| RAS inhibitor |
Reduction of Inflammation |
| Glucocorticoid |
Reduction of Inflammation |
| CRISPR-Cas13d |
Target cleavage of the Viral RNA genome using AAVs as delivery vehicle |
The most promising development in antiviral development for COVID-19 will likely come from the clinical trials of Remdisivir, an antiviral nucleotide analog in China [63]. Gilead, the company responsible for developing the drug, has reached a deal with the Chinese Health Bureau in order to accelerate through clinical trials. If Remdisivir can be approved for use soon, it would likely save tens of thousands of lives before a vaccine for COVID-19 can be developed.
Chloroquine is an inexpensive and reliable anti-malarial that has been used for more than 70 years [22]. Due to the urgent demand for finding treatments for COVID-19, chloroquine or chloroquine phosphate is recommended to treat COVID-19, especially for its associated pneumonia.
In the past, glucocorticoids have been used to treat other coronaviruses by reducing inflammation. This treatment helped to reduce mortality in SARS epidemic. Due to its similar genome, it has been recommended to use glucocorticoids to treat SARSCoV- 2. However, usage of glucocorticoids should be strictly controlled [84].
Monoclonal antibodies are being developed to target the viral surface proteins, such as the Spike (S). Other small molecule drugs are being investigated on their ability to target viral proteases, polymerases and other key enzymes [20].
In terms of new molecular treatment approaches, Nguyen et al. [40] have suggested using a CRISPR/Cas13d system to directly target the viral RNA genome without any effects on the human genome. Interestingly, they suggested using an adeno-associated virus (AAV) as a drug delivery vehicle to specifically target lung tissue, minimizing other side effects.
Nutritional supplements should also be included in the treatment of COVID-19. Vitamins A, B, C, D, and E all enhance the immune system in some capacity and can target many types of viruses such as the SARS-CoV-1, MERS-CoV, and avian coronavirus [80]. Certain vitamins might be effective for bodily defense against SARS-CoV-2. Omega-3 polyunsaturated fatty acids can mediate inflammation. Selenium can act as a coenzyme to assist antioxidants in preventing free radical formation. Zinc can help maintain and develop immune cells, while a deficiency in iron can impair host immunity [80].
For patients that have refractory hypoxemia, extracorporeal membrane oxygenation (ECMO) is a viable treatment. ECMO is a cardiopulmonary bypass where the patient’s venous blood is removed from the body and pumped through an artificial membrane lung [37]. However, ECMO is a finite resource and in large outbreaks such as COVID-19, only the most critically ill patients should receive ECMO therapy [37].
In terms of pediatric guidelines, the general treatment plan is bed rest and supportive treatments; ensuring enough calorie and water intake; maintaining water electrolyte balance and homeostasis. Antiviral therapies such as Interferon- α2b nebulization and Lopinavir can also be used. However, antibiotic applications and immunomodulating therapy should be avoided [12].
Prevention
In hospital settings, SARS-CoV-2 can easily spread and cross infect patients and healthcare providers. In these nosocomial infections, oral barriers are important in preventing the virus from further spreading. During oral examination, medical staff are in close contact with the patients who are sick. A cough or sneeze from such patient would allow the virus to escape the host via saliva or blood droplets; nebulized secretions, saliva, and blood from small aerosol particles suspended in the air [31].If there are no protective measures, large particles of droplets can directly spread and contaminate the conjunctiva of the eyes, the nose, and the mouth mucosa of other patients and medical staff leading to an infection [81].
To prevent SARS-CoV-2 from further spreading, countermeasures can be placed in hospitals to ensure that the virus does not cross-infect. Sterilization techniques, waste management and ultraviolet radiation all should be implemented. Sterilization techniques must be strictly implemented with the use of concentrated chlorine solution. After the oral diagnosis and treatment of suspected or confirmed patients, waste should be placed in double-layer anti-leakage special packaging bags for careful disposal [31].
The SARS-CoV-2 is sensitive to ultraviolet light and heat. To effectively inactivate the virus, a temperature of at least 56º C is required with the addition of solvents such as ether, 75% ethanol, chlorine-containing disinfectant, peroxyacetic acid, or chloroform [81]. It is also worth mentioning that Kampf et al. [24] has shown that SARS-CoV-2 can persist on metal, plastic or glass surfaces for up to 9 days, thus warranting frequent sanitations of surfaces that are in proximity with patients or potential carriers.
Secondary protection should be worn by all medical staff treating patients with fever-like symptoms and tertiary protection should be used when patients are suspected or confirmed with an infection [76]. A crucial component of this level of protection is the N95 respirator face mask. China as well as other nations have been ramping up productions in response to the urgent shortage of such supplies. Special care is often needed to protect ophthalmologists as well as head and neck surgical staff, who are often at higher risk of exposure [76,81].
A great challenge that Chinese medical personnel has faced in this first phase of fighting COVID-19 is the shortage of barrier devices, including N95 face masks and other protective gear. In response to this, various communities, domestic and international have collaborated in donating needed medical supplies to the “frontlines”. China has also ramped up its production of medical supplies to meet the rising demand [65].
Additional protection may be needed to address recent findings that SARS-CoV-2 may be infective through the oral-fecal route [79]. Special treatment of patients’ fecal excretions may be needed as to prevent further spread of the virus through the common sewage systems. Personnel involved in the sanitation and management of sewage should be given additional training and protection.
Regional blockades should be used in early stages of the infection to prevent the spread from areas of high case counts [55]. This strategy has already been implemented in multiple Chinese cities, most notably Wuhan, the origin of the epidemic. Airports need to take greater responsibility in screening passengers during this crucial time. Quilty et al. [46] estimated that 46% of infected passengers would not be effectively detected under standard airport protocols (p<0.05).This presents a need for airports to redesign and improve their screening protocols in response for this new viral threat.
The ultimate form of prevention we hope to achieve would be through the eventual creation of a vaccine. Extensive molecular and genetic research is currently underway for COVID-19, where several key viral sites and regions have been successfully identified. Therapeutic targeting of such epitopes may help researchers advance their search for a vaccine or an effective treatment [5]. Specific molecular findings of SARS-CoV-2 and how they lead to pathogenesis will be discussed in following sections.
It is important to note that patients that are cured of their infections are still considered at-risk for re-infection as there has yet to be extensive research on acquired immunity for SARSCoV- 2. Further studies should be performed to ascertain their level of risk compared to the general population as it may be increased due to their compromised immune system.
Pathogenicity
The SARS-CoV-2, SARS-CoV-1, and MERS-CoV show several similarities regarding the clinical presentations, which can vary from asymptomatic infection to severe disease. After the successful inoculation of the SARS-CoV-2 with surface layers of human airway epithelial cells, the host cell will experience a cytopathic effect leading to a cessation of cilium beating of the cells. The SARS-CoV-2 infection has a cluster of onsets, with an increased risk for older males with comorbidities, and can result in severe and even fatal respiratory diseases. The major clinical symptoms resulting from SARS-CoV-2 infection at the prodromal phase include fever, dry cough, myalgia, fatigue, and diarrhea [34]. Currently, the SARS-CoV-2 is considered less pathogenic than SARS-CoV and MERS-CoV, but further studies are needed to ascertain this [11]. It can be noted that SARS-CoV-2 case of fatality rate is 3% and R0 is between 1.4-5.5 while both SARSCoV- 1 and MERS have higher fatality rate and R0. An increased R0 value indicates an increase in virulence as mentioned in a previous section, transmissibility [9]. It should be noted that these are just preliminary estimates and may be higher in other regions affected by the virus.
One potential caveat when considering the geographical discrepancies in SARS-CoV-2 pathogenicity is patients’ prior exposure to other coronaviruses. Antibody-Dependent Enhancement (ADE) of the SARS-CoV-2 may be a factor at play when considering the greater proportion of severe cases in Hubei Province, China compared to that of the rest of the world. ADE is a result of prior exposures to similar viral epitopes and may result in chronic inflammation and lymphopenia that may contribute to poorer outcomes of certain patients [58].
Molecular Characteristics
The Coronavirus (CoV) is a single stranded, positive-sense RNA virus. The current classification for CoVs is that they belong to the realm Riboviria, order Nidovirales, suborder Cornidovirineae, family Coronaviridae and subfamily Coronavirinae [1]. Knowing that the coronaviruses belong to the order Nidovirales allow scientists to better understand their behavioral properties of replicase or transcriptase. Coronaviruses belonging to subfamily Coronavirinae mainly cause respiratory and gastrointestinal tract infection and can be genetically grouped into four major genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. The former two genera, Alphacoronavirus and Betacoronavirus, primarily infect mammals whereas the latter two, Gammacoronavirus and Deltacoronavirus, primarily infect birds. Understanding the origin for these pathogens is crucial in characterizing the viruses’ behavior and pathogenicity [1].
Before the COVID-19 pandemic, only six species of human CoVs have been identified. Two of the six species, known as HCoVNL63 and HCoV-229E, belong to the genus Alphacoronavirus. The genus Betacoronavirus is comprised of the remaining four species known as HCoV-OC43, HCoV-HKU1, severe acute respiratory syndrome coronavirus (SARS-CoV-1), and Middle East respiratory syndrome coronavirus (MERS-CoV) [71]. Despite being discovered in the mid-1960s, the Coronavirus did not receive its world-wide recognition until the 2003 SARS pandemic, followed by the 2012 MERS, and most recently the SARS-CoV-2 (2019 nCoV) outbreak. SARS-CoV-1 originated in bats and spread to civets and later to humans. MERS-CoV originated in bats and spread to camels and later to humans. SARS-CoV-1 and MERS-CoV are both pathogenic and understanding their underlying molecular basis will help give insight to the SARS-CoV-2’s mechanism of infection, pathology, and treatments [72]. The genetic sequence analysis revealed that the SARS-CoV-2 belongs to the β-coronavirus genus, with a 79.0% nucleotide identity to SARS-CoV-1 and 51.8% identity to MERSCoV. Furthermore, it has been reported that SARS-CoV-2 is 96% identical across the entire genome to a bat coronavirus. Table 4 shows the open reading frames and the subunits of SARS-CoV-2.
Table 4 Open reading frames, subunits, and their functions for Coronaviruses.
| Open Reading Frames (ORFs) |
Subunits of Coronaviruses [11] |
Functions |
ORF 1
***ORF 1 contains codes for 67% of CoV genome |
nsp 1 |
Cellular mRNA degradation, inhibiting IFN signaling |
| nsp 2 |
Unknown |
| nsp 3 |
PLP, polypeptides cleaving, blocking host innate immune response, promoting cytokine expression |
| nsp 4 |
DMC formation |
| nsp 5 |
3CLpro, Mpro, polypeptides cleaving, inhibiting IFN signaling |
| nsp 6 |
Restricting autophagosome expansion, DMV formation |
| nsp 7 |
Cofactor with nsp8 and nsp12 |
| nsp 8 |
Cofactor with nsp7 and nsp12, primase |
| |
nsp 9 |
Dimerization and RNA binding |
| nsp 10 |
Scaffold protein for nsp14 and nsp16 |
| nsp 11 |
Unknown |
| nsp 12 |
Primer dependent RdRp |
| nsp 13 |
RNA helicase, 5′ triphosphatase |
| nsp 14 |
Exoribonuclease, N7?MTase |
| nsp 15 |
Endoribonuclease, evasion of dsRNA sensors |
| nsp 16 |
2′?O?MTase; avoiding MDA5 recognition, negatively regulating innate immunity |
| Other ORFs |
S protein |
spikes on the viral surface and they are responsible for attachment to host receptors |
| E protein |
plays a role in virus assembly and release, and it involved in viral pathogenesis |
| M protein |
has three transmembrane domains and it shapes the virions, promotes membrane curvature, and binds to the nucleocapsid |
| N protein |
contains two domains, can bind virus RNA genome via different mechanisms. It is reported that N protein can bind to nsp3 protein to help tether the genome to RTC, and package the encapsulated genome into virions |
In general, the coronaviruses possess a genome size of approximately 26,000-32,000 bases, including a variable number of translatable codons or open reading frames (ORFS) of 6-11 [70]. The first ORF contains approximately 67% of the entire genome which encodes for 16 non-structural proteins (NSPs), while the remaining ORFs encode for accessory proteins and structural proteins. The four major structural proteins are the spike surface glycoprotein (S), small envelope protein (E), matrix protein (M), and nucleocapsid protein (N). The spike surface glycoprotein allows the virus to bind to receptors, and therefore determines the virus’s preferred range of hosts, also referred to as its host tropism. This mechanism is used by both SARS- CoV-1 and MERS-CoV to infect the host. With their specific spike surface glycoproteins, CoVs can bind to the host receptor. For example, the spike glycoprotein of SARS-CoV-1 binds to angiotensinconverting enzyme 2 (ACE2) as one of the main receptors, whereas MERS-CoV uses dipeptidyl peptidase 4 (DPP4 or CD26) as one of the main receptors. With this insight, analysis suggests that SARS-CoV-2 has close evolutionary association with SARSCoV- 1, therefore, the mechanism of SARS-CoV-2 can be better understood by understanding the mechanism of SARS-CoV-1 in Figure 1.
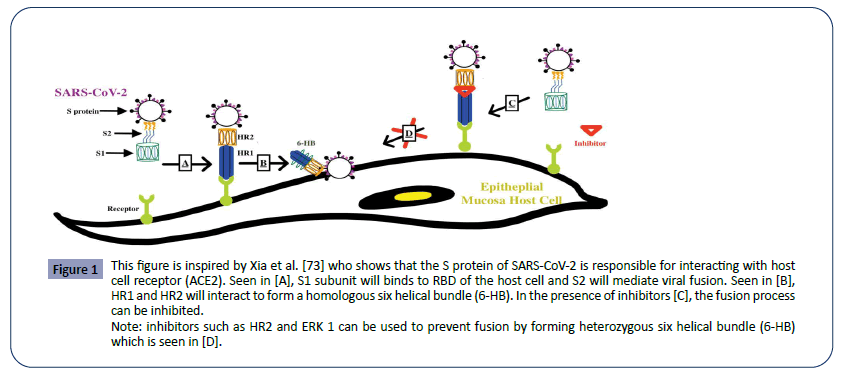
Figure 1: This figure is inspired by Xia et al. [73] who shows that the S protein of SARS-CoV-2 is responsible for interacting with host cell receptor (ACE2). Seen in [A], S1 subunit will binds to RBD of the host cell and S2 will mediate viral fusion. Seen in [B], HR1 and HR2 will interact to form a homologous six helical bundle (6-HB). In the presence of inhibitors [C], the fusion process can be inhibited.
SARS-CoV-1 spike protein has two subunits, S1 and S2. The S1 subunit binds to the ACE2 receptor on the host cells. The S2 subunit plays a key role in mediating viral fusion with and entry into the host cell [73]. To accomplish this, SARS-CoV has a heptad repeat 1 (HR1) and a heptad repeat 2 (HR2) which can interact and form a homologous six helical bundle (6-HB). This helps bring the viruses closer to the host cells and facilitate the fusion process. Thus, by inhibiting the formation of viral homologous 6-HB, fusion between the host cell and virus can be prevented. Some examples of these inhibitors are HR2P and EK1 [73].
In the SARS-CoV-2, a similar mechanism of infection can be observed via its highly conserved spike protein subunits. Again, The S1 subunit will facilitate the viral fusion by binding to the host cell receptor, ACE2. S1 subunit can further be divided into the N-terminal domain and C-terminal RBD domain that directly interact with the host receptor. Although both spike proteins bind to the same host receptor, SARS-1CoV-1 and SARS-1CoV-2 have subtle differences in their spike protein. Residues at position 442, 472, 479, 487, and 491 on the SARS-CoV-1 S-protein are present at the receptor complex interface and critical for cross-species and human-to-human transmission. Despite the highly conserved regions in the RBD domain of the SARS-CoV-2 S-protein, four of the five critical residues are not preserved except for Tyr491. Thus, some studies even suggest that SARS-CoV-2 may bind to a different receptor than ACE2. And even if it did bind ACE2, the effects of this interaction in facilitating human transmission need to be further considered [77]. Note that MERS-CoV S-protein displayed very little homology compared to the SAR-CoV-1 S-protein; MERS-CoV S-protein does not bind to ACE2 receptor, instead interacting with the human dipeptidyl peptidase 4 (DPP4). It should be noted that the furin-like site identified in the S protein of SARS-CoV-2 is unique in beta coronaviruses. Furinlike sites have been identified in other coronaviruses before but have have not been discovered in the beta-lineage that the SARSCoV- 2 derives from Coutard et al. [15].
Specific to the RBD domain, SARS-CoV-2 RBD domain has a distinct loop. Unlike other CoVs, like SARS-CoV-1, RBD domain of SARSCoV- 2 replaces rigid prolyl residues with flexible glycyl residues and has a unique phenylalanine F468 in the flexible loops which provides hydrophobic interaction with the ACE2 motif. These specific mutations enhance SARS-CoV-2 RBD binding affinity with ACE2 [11].
Another notable feature that sets SARS-CoV-2 apart from its viral family members, such as the SARS-CoV-1, is the presence of a RRAR (Arginine-Arginine-Alanine-Arginine) motif at the border of the S1/S2 proteins. This sequence was also flanked by several O-linked-glycosylation sites that are thought to protect this important motif [68]. The detailed function of this RRAR motif has not yet been determined, but it is believed to facilitate cell-cell fusion through its abundant basic residues, without necessarily affecting mechanisms of viral entry [27].
Due to mutations in the RBD domain of SARS-CoV-2, hydrogen bond interactions have been lost due to the replacement of Arg426 with Asn426 in the S-protein. This change increased the free binding energy for SARS-CoV-2 S-protein by 28 kcal/ mol when compared to SARS-CoV-1 S-protein binding. Despite this finding, the SARS-CoV-2 S-protein is still regarded to have a stronger binding interaction to human ACE2. Thus, even with four of five critical residues replaced, SARS-CoV-2 S-protein was found to have a significant binding affinity to ACE2 molecule. This phenomenon could potentially be explained by the unaltered structural confirmation of SARS-CoV-2 S-protein. It was demonstrated that the SARS-CoV-2 S-protein and SARS-CoV-1 S-protein shared an almost identical 3-D structure in the RBD domain, thus maintaining similar van der Waals and electrostatic properties in the interaction interface.
Recent genetic analysis on 86 complete/near-complete genomes of SARS-CoV-2 was performed by Phan et al. [44] and various mutations were found in coding as well as non-coding regions. This may suggest that the virus has great genetic diversity and is capable of further evolutions that may complicate the process of finding treatments and developing a vaccine.
Miscellaneous Influences
Additionally, it is important to discuss the significant influence of COVID-19 in broader, non-medical contexts and settings. Ayittey et al. [2] predicts there will be interrupted global trade and supply chains that force multinational business to make hard decisions on their business plans. At the same time, the asset prices have depressed. With the limited information businesses have about COVID-19, it is vital for businesses to keep up to date with the virus and to plan for future business plans. Shanghai Composite index fell 7.7% since the outbreak. China is expected to lose $62 billion in revenue in the first quarter along, while the rest of the world will lose $280 billion in that same time period [2].
Human virologists are not the only ones studying coronaviruses, veterinary virologists have also performed research on them extensively. Their suggestion to preventing viral zoonosis is to maintain barriers between animal natural reservoirs and human society [36]. In other words, a public health barrier needs to be in place between potential viral animal hosts and human society. As human activity continues to expand, such a barrier has become more difficult to create and maintain.
There have been recent conspiracy theories suggesting that SARS-CoV-2 does not have a natural origin, and instead was an artificial creation [6]. However, genomic evidence has shown that SARS-CoV-2 likely originated in a wildlife source before it evolved to gain access to human ACE2 receptors [6]. There is no definitive proof of this. More studies are needed to ascertain the origins of the virus. Calisher et al. [6] urges researchers to collaborate in a global fight against the virus and to reject misinformation and false claims.
With the onset of this new virus, patients and healthcare providers may experience fear and uncertainty. According to Xiang et al. [75] the solution to this is to have mental health teams provide strong and lasting support for patients and healthcare workers. This includes clear communication about any updates on the outbreak to health institutions, sincere and consistent communication between family members and the patient in isolation, as well as regular clinical screenings for signs of depression [74].
Finally, with the recent outbreak of COVID-19, it may be the opportune moment to utilize Augmented Intelligence (AI) to help predict viral transmission [35]. There have already been some AI that have been used to map potential spreads of COVID-19, such as HealthMap ™ and BlueDot ™. Both AI programs have previously proven successful in prediction of international spread of viruses, such as the 2015 Zika virus in Brazil [35]. AI can eventually help differentiate between the new viral infections, such as COVID-19, and other more benign causes of respiratory illnesses [35].
Even though molecular testing and research for COVID-19 was implemented quickly in the European Union and other developing nations as part of this global response, it is important to note that current tests are far from ideal in validating clinical specificity and sensitivity of implemented tests against SARS-CoV-2 [47]. Current barriers include the limited availability of primers/probes, positive controls, and shortages of required personnel [47].
Conclusion
Three months in to the COVID-19 outbreak, physicians, researchers and people of all backgrounds have worked tirelessly together to advance our collective knowledge of this new menace threatening public health on a global scale.
Basic transmission characteristics have been ascertained and an initial diagnostic protocol have been established using qRT-PCR. Notably, there has been a push to also establish a radiological diagnostic protocol using HRCT, especially after a study demonstrating HRCT achieving a higher diagnostic sensitivity than qRT-PCR. Some patients that tested negative on qRT-PCR were successfully identified using HRCT. A study on viral loads of asymptomatic vs. symptomatic patients revealed that asymptomatic patients are potentially equally likely to contribute to transmission.
Deep dives into the molecular and genetic composition of SARS-CoV-2 revealed some of its key similarities and differences compared toto other coronaviruses. Notably, a distinctly structured Spike (S) protein on the surface of SARS-CoV-2 may contribute to its strong transmission mechanisms via interactions with human respiratory tract receptors, likely ACE2. A subtle yet unique RRAR motif found in the transitional border zone of S1/S2 proteins may facilitate cell-cell fusion Other amino acid substitutions in multiple positions may have also resulted in the SARS-CoV-2 having stronger transmission capabilities, yet decreased pathogenicity compared to its other viral family members.
Such discoveries made in the microscopic features of the SARS-CoV-2 may ultimately translate to great advances in the development of treatments and vaccines. Although current treatment plans for COVID-19 consist of mostly supportive and nutritional therapy, multiple anti-viral drugs have entered clinical trial, such as Remdisivir. Although current treatment plans for COVID-19 consist of mostly supportive and nutritional therapy, multiple anti-viral drugs have entered clinical trial, such as Remdisivir. Other treatment methods employ molecular techniques, such as Ras-Inhibition or the CRISPR-Cas13d system, delivered via an adeno-associated virus (AAV). Clinicians and researchers are seeking innovative solutions to treating this new health threat every day.
Ninety days in, we may still not know as much as we would like about the SARS-CoV-2. But as new information is gathered around the world every day, our understanding will only grow stronger. This review hopes to capture the significant findings of this initial period of the COVID-19 outbreak and inspire more exploration into various aspects of the virus that may contribute to the resolution of the crisis.
Acknowledgements
We thank Dr. Robert F. Garry Jr. of the Department of Microbiology and Immunology at Tulane University School of Medicine for his expertise in reviewing and editing this paper. We pay our respects to all the clinicians, nurses and medical workers on the frontlines of fighting the virus.
Declaration of Interests
There are no conflicting interests to declare for the authors of this paper.
Funding Details
No funding was received for the research performed in this paper.
26659
References
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, et al. (2020) Severe Acute Respiratory Syndrome-related coronavirus: The species and its viruses- a statement of the coronavirus study group. bioRxiv.
- Ayittey FK, Ayittey MK, Chiwero NB, Kamasah JS, Dzuvor C (2020) Economic Impacts of Wuhan 2019-nCoV on China and the World. J Med Virol 92.
- Backer JA, Klinkenberg D, Wallinga J (2020) Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20-28 January 2020. Euro Surveill 25.
- Bai Y, Yao L, Wei T, Tian F, Jin DY, et al. (2020) Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA.
- Baruah V, Bose S (2020) Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J Med Virol 92: 495-500.
- Calisher C, Carroll D, Colwell R, Corley RB, Daszak P (2020) Statement in support of the scientists, public health professionals, and medical professionals of China combatting COVID-19. The Lancet 395: PE42-E43.
- Chan JF, Yuan S, Kok KH, To KK, Chu H, et al. (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395: 514-523.
- Channappanavar R, Fett C, Mack M, Ten Eyck PP, Meyerholz DK (2017) Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection. J Immunol 198: 4046-4053.
- Chen L, Liu W, Zhang Q, Xu K, Ye G, et al. (2020) RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect 9: 313-319.
- Chen N, Zhou M, Dong X, Qu J, Gong F, et al. (2020) Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395: 507-513.
- Chen Y, Guo Y, Pan Y, Zhao ZJ (2020) Structure analysis of the receptor binding of 2019-nCoV. Biochem Biophys Res Commun.
- Chen ZM, Fu JF, Shu Q (2020) New coronavirus: new challenges for pediatricians. World J Pediatr.
- Chen ZM, Fu JF, Shu Q, Chen YH, Hua CZ, et al. (2020) Diagnosis and treatment recommendations for pediatric respiratory infection caused by the 2019 novel coronavirus. World J Pediatr.
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, et al. (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25: 2000045.
- Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, et al. (2020) The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176: 104742.
- Dennis Lo YM, Chiu RWK (2020) Racing towards the development of diagnostics for a novel coronavirus (2019-nCoV). Clin Chem 66: 503-504.
- Du B, Qiu HB, Zhan X, Wang YS, Kang HYJ, et al. (2020) Pharmacotherapeutics for the New Coronavirus Pneumonia. Zhonghua Jie He He Hu Xi Za Zhi 43: E012.
- Du Z, Wang L, Cauchemez S, Xu X, Wang X, et al. (2020) Risk for Transportation of 2019 Novel Coronavirus Disease from Wuhan to Other Cities in China. Emerg Infect Dis 26.
- Fang Y, Zhang H, Xie J, Lin M, Ying L, et al. (2020) Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology 200432.
- Fauci AS, Lane HC, Redfield RR (2020) Covid-19 - Navigating the Uncharted. N Engl J Med.
- Gallego V, Nishiura H, Sah R, Rodriguez-Morales AJ (2020) The COVID-19 outbreak and implications for the Tokyo 2020 Summer Olympic Games. Travel Med Infect Dis 101604.
- Gao J, Tian Z, Yang X (2020) Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends 14:72-73.
- Gao QY, Chen YX, Fang JY (2020) 2019 novel coronavirus infection and gastrointestinal tract. J Dig Dis.
- Kampf G, Todt D, Pfaender S, Steinmann E (2020) Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect 104: 246–251.
- Kim JY, Ko JH, Kim Y, Kim YJ, Kim JM, et al. (2020) Viral Load Kinetics of SARS-CoV-2 Infection in First Two Patients in Korea. J Korean Med Sci 35: e86.
- Korean Society of Infectious Diseases , Korean Society of Pediatric Infectious Diseases , Korean Society of Epidemiology , Korean Society for Antimicrobial Therapy , Korean Society for Healthcare-associated Infection Control and Prevention, et al. (2020) Report on the Epidemiological Features of Coronavirus Disease 2019 (COVID-19) Outbreak in the Republic of Korea from January 19 to March 2, 2020. J Korean Med Sci 35: e112.
- Anderson KG, Rambaut A, Lipkin WL, Holmes EC, Garry RF (2020) The Proximal Origin of SARS-CoV-2.
- Li H, Wang YM, Xu JY, Cao B (2020) Potential antiviral therapeutics for 2019 Novel Coronavirus. Zhonghua Jie He He Hu Xi Za Zhi 43: E002.
- Li J, Li J, Xie X, Cai X, Huang J, et al. (2020) Game consumption and the 2019 novel coronavirus. Lancet Infect Dis 20: P275-276.
- Li Q, Guan X, Wu P, Wang X, Zhou L, et al. (2020) Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med.
- Li ZY, Meng LY (2020) The prevention and control of a new coronavirus infection in department of stomatology. Zhonghua Kou Qiang Yi Xue Za Zhi 55: E001.
- Lim J, Jeon S, Shin HY, Kim MJ, Seong YM, et al. (2020) Case of the Index Patient Who Caused Tertiary Transmission of COVID-19 Infection in Korea: the Application of Lopinavir/Ritonavir for the Treatment of COVID-19 Infected Pneumonia Monitored by Quantitative RT-PCR. J Korean Med Sci 35: e79.
- Lipsitch M, Swerdlow DL, Finelli L (2020) Defining the Epidemiology of Covid-19 - Studies Needed. N Engl J Med 382: 1194-1196.
- Liu Y, Gayle AA, Wilder-Smith A, Rocklov J (2020) The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med 27.
- Long JB, Ehrenfeld JM (2020) The Role of Augmented Intelligence (AI) in Detecting and Preventing the Spread of Novel Coronavirus. J Med Syst 44: 59.
- Lorusso A, Calistri P, Petrini A, Savini G, Decaro N (2020) Novel coronavirus (SARS-CoV-2) epidemic: a veterinary perspective. Vet Ital.
- MacLaren G, Fisher D, Brodie D (2020) Preparing for the Most Critically Ill Patients With COVID-19: The Potential Role of Extracorporeal Membrane Oxygenation. JAMA.
- Mahase E (2020) Covid-19: WHO declares pandemic because of "alarming levels" of spread, severity, and inaction. BMJ 368: m1036.
- Mandl JN, Schneider C, Schneider DS, Baker ML (2018) Going to Bat(s) for Studies of Disease Tolerance. Front Immunol 9: 2112.
- Nguyen TM, Zhang Y, Pandolfi PP (2020) Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res.
- Pan F, Ye T, Sun P, Gui S, Liang B, et al. (2020) Time Course of Lung Changes On Chest CT During Recovery From 2019 Novel Coronavirus (COVID-19) Pneumonia. Radiology 200370.
- Pan X, Chen D, Xia Y, Wu X, Li T, et al. (2020) Asymptomatic cases in a family cluster with SARS-CoV-2 infection. Lancet Infect Dis.
- Pan Y, Guan H, Zhou S, Wang Y, Li Q, et al. (2020) Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur Radiol.
- Phan T (2020) Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol 104260.
- Centers for Disease Control and Prevention (2020) Transcript for CDC Media Telebriefing: Update on COVID-19.
- Quilty BJ, Clifford S, Cmmid nCo VWG, Flasche S, Eggo RM (2020) Effectiveness of airport screening at detecting travellers infected with novel coronavirus (2019-nCoV). Euro Surveill 25.
- Reusken C, Broberg EK, Haagmans B, Meijer A, Corman VM, et al. (2020) Laboratory readiness and response for novel coronavirus (2019-nCoV) in expert laboratories in 30 EU/EEA countries, January 2020. Euro Surveill 25.
- Rodriguez-Morales AJ, Bonilla-Aldana D, Balbin-Ramon GJ, Rabaan AA, Sah R, et al. (2020) History is repeating itself: Probable zoonotic spillover as the cause of the 2019 novel Coronavirus Epidemic. Infez Med 28: 3-5.
- Ronco C, Navalesi P, Vincent JL (2020) Coronavirus epidemic: preparing for extracorporeal organ support in intensive care. Lancet Respir Med 8: P240-241.
- Shi H, Han X, Zheng C (2020) Evolution of CT Manifestations in a Patient Recovered from 2019 Novel Coronavirus (2019-nCoV) Pneumonia in Wuhan, China. Radiology 295: 18.
- Shigemura J, Ursano RJ, Morganstein JC, Kurosawa M, Benedek DM (2020) Public responses to the novel 2019 coronavirus (2019-nCoV) in Japan: Mental health consequences and target populations. Psychiatry Clin Neurosci.
- Smith N, Fraser M (2020) Straining the System: Novel Coronavirus (COVID-19) and Preparedness for Concomitant Disasters. Am J Public Health pp: e1-e2.
- Sohrabi C, Alsafi Z, O'Neill N, Khan M, Kerwan A, et al. (2020) World Health Organization declares Global Emergency: A review of the 2019 Novel Coronavirus (COVID-19). Int J Surg 76: 71-76.
- Special Expert Group for Control of the Epidemic of Novel Coronavirus Pneumonia of the Chinese Preventive Medicine Association (2020) An update on the epidemiological characteristics of novel coronavirus pneumoniaCOVID-19. Zhonghua Liu Xing Bing Xue Za Zhi 41: 139-144.
- Strategy and Policy Working Group for NCIP Epidemic Response (2020) Urgent research agenda for the novel coronavirus epidemic: transmission and non-pharmaceutical mitigation strategies. Zhonghua Liu Xing Bing Xue Za Zhi 41: 1-6.
- Sun ML, Yang JM, Sun YP, Su GH (2020) Inhibitors of RAS Might Be a Good Choice for the Therapy of COVID-19 Pneumonia. Zhonghua Jie He He Hu Xi Za Zhi 43: E014.
- COVID-19 National Incident Room Surveillance Team (2020) COVID-19, Australia: Epidemiology Report 2 (Reporting week ending 19:00 AEDT 8 February 2020). Commun Dis Intell (2018) 44.
- Tetro JA (2020) Is COVID-19 Receiving ADE From Other Coronaviruses? Microbes Infect.
- To KK, Tsang OT, Chik-Yan YC, Chan KH, Wu TC, et al. (2020) Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis.
- Velavan TP, Meyer CG (2020) The Covid-19 epidemic. Trop Med Int Health 25: 278-280.
- Wang D, Hu B, Hu C, Zhu F, Liu X, et al. (2020) Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA.
- Wang M, Cao R, Zhang L, Yang X, Liu J, et al. (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30: 269-271.
- Wang S, Zhang Y, Lui S, Peng H, Mackey V, et al. (2020) Coronaviruses and the Associated Potential Therapeutics for the Viral Infections. Journal of Infectious Diseases and Therapy 8: 2.
- Wang W, Tang J, Wei F (2020) Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J Med Virol 92: 441-447.
- Wang X, Zhang X, He J (2020) Challenges to the system of reserve medical supplies for public health emergencies: reflections on the outbreak of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic in China. Biosci Trends 14: 3-8.
- Wang Y, Kang H, Liu X, Tong Z (2020) Combination of RT-qPCR Testing and Clinical Features For Diagnosis of COVID-19 facilitates management of SARS-CoV-2 Outbreak. J Med Virol.
- Wassenaar TM, Zou Y (2020) 2019_nCoV: Rapid classification of betacoronaviruses and identification of traditional Chinese medicine as potential origin of zoonotic coronaviruses. Lett Appl Microbiol.
- Wei M, Yuan J, Liu Y, Fu T, Yu X, et al. (2020) Novel Coronavirus Infection in Hospitalized Infants Under 1 Year of Age in China. JAMA.
- Working Group of Novel Coronavirus, PUMCH (2020) Diagnosis and clinical management of 2019 novel coronavirus infection: an operational recommendation of Peking Union Medical College Hospital (V2.0). Zhonghua Nei Ke Za Zhi 59: 186-188.
- Wu A, Peng Y, Huang B, Ding X, Wang X, et al. (2020) Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 27: 325-328.
- Wu JT, Leung K, Leung GM (2020) Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet 395: P689-697.
- Wu Y (2020) Compensation of ACE2 Function for Possible Clinical Management of 2019-nCoV-Induced Acute Lung Injury. Virol Sin.
- Xia S, Zhu Y, Liu M, Lan Q, Xu W, et al. (2020) Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol.
- Xiang YT, Yang Y, Li W, Zhang L, Zhang Q, et al. (2020) Timely mental health care for the 2019 novel coronavirus outbreak is urgently needed. Lancet Psychiatry 7: P228-229.
- Xie X, Zhong Z, Zhao W, Zheng C, Wang F, et al. (2020) Chest CT for Typical 2019-nCoV Pneumonia: Relationship to Negative RT-PCR Testing. Radiology 200343.
- Xu K, Lai XQ, Liu Z (2020) Suggestions for prevention of 2019 novel coronavirus infection in otolaryngology head and neck surgery medical staff. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 55: E001.
- Xu X, Yu C, Zhang L, Luo L, Liu J (2020) Imaging features of 2019 novel coronavirus pneumonia. Eur J Nucl Med Mol Imaging 47: 1275-1280.
- Xu Z, Shi L, Wang Y, Zhang J, Huang L, et al. (2020) Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 8: P420-422.
- Yeo C, Kaushal S, Yeo D (2020) Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol 5: P335-337.
- Zhang L, Liu Y (2020) Potential Interventions for Novel Coronavirus in China: A Systemic Review. J Med Virol 92: 479-490.
- Zhang MC, Xie HT, Xu KK, Cao Y (2020) Suggestions for disinfection of ophthalmic examination equipment and protection of ophthalmologist against 2019 novel coronavirus infection. Zhonghua Yan Ke Za Zhi 56: E001.
- Zhang MQ, Wang XH, Chen YL, Zhao KL, Cai YQ, et al. (2020) Clinical features of 2019 novel coronavirus pneumonia in the early stage from a fever clinic in Beijing. Zhonghua Jie He He Hu Xi Za Zhi 43: E013.
- Zhang W, Du RH, Li B, Zheng XS, Yang XL, et al. (2020) Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect 9: 386-389.
- Zhao JP, Hu Y, Du RH, Chen ZS, Jin Y, et al. (2020) Expert consensus on the use of corticosteroid in patients with 2019-nCoV pneumonia. Zhonghua Jie He He Hu Xi Za Zhi 43: E007.
- Zhao S, Lin Q, Ran J, Musa SS, Yang G, et al. (2020) Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int J Infect Dis 92: 214-217.
- Zhou T, Liu Q, Yang Z, Liao J, Yang K, et al. (2020) Preliminary prediction of the basic reproduction number of the Wuhan novel coronavirus 2019-nCoV. J Evid Based Med 13: 3-7.
- Zou L, Ruan F, Huang M, Liang L, Huang H, et al. (2020) SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med 382: 1177-1179.