Keywords
Shewanella, Tilapia, Bal?k, Çiftlik, RAPD
Giri?
The organism now known as Shewanella pu-trefaciens has undergone an extensive taxonomic evolution during the past seven decades. The or-ganism was first isolated from tainted butter by Derby and Hammer (1931) and classified as a member of the genus Achromobacter which is no longer extant. It was later transferred to the ge-nus Pseudomonas by Long and Hammer (1941). It was then allocated to the genus Alteromonas by Lee et al (1977) on the basis of its much lower mols% G+C DNA content than the acceptable range of 58 to 65 mols% G+C for members of the genus Pseudomonas. It was transferred by Mac-Donell and Colwell (1985) to the newly establish genus Shewanella under the family Vibrionaceae due to its perceived closer relationship with the genus Vibrio on the basis of 5S rRNA sequence data.
Bacterial isolates designated as S. putrefaciens are derived from a wide variety of sources in-cluding environmental samples: soil, oil fields, sea water, refrigerated foods such as fish, poultry, beef, and human clinical infections. Levin (1972) was the first to address the question as to whether the intense refrigerated food spoilage isolates of P. putrefaciens were identical to clini-cal isolates.
Castell et al. (1949) were the first to report that P. putrefaciens was involved in the spoilage of marine fish. Chai et al. (1968) found that the initial population of S. putrefaciens on fresh had-dock fillets was consistently below 4% and that during refrigerated storage increased to 50 to 90% of the total bacterial population at the time of spoilage. Jørgensen and Huss (1989) con-cluded that S. putrefaciens constitutes the main spoilage organism on fish stored at 0oC, irrespec-tive of the origin of the fish and whether the fish is vacuum packed or stored in ice. Gram (1992; 1996) similarly stated S. putrefaciens to be the most important bacterium during spoilage of iced marine fish. The organism is also one of the pre-dominant spoilage organisms of refrigerated meat (Borch et al. 1996).
Numerous reports have implicated S. putrefa-ciens as the cause of various human infections such as cellulitis (Chen et al. 1997), peritonitis (Dan et al. 1992), abscesses (Yohe et al. 1997; Pagani et al. 2003), bacteremia (Brink et al. 1995; Pagani et al. 2003), and ear infections (von Graevenitz and Simon, 1970; Holmes et al. 1975). Patients developing bacteremia have been found to have an underlying illness such as di-abetes mellitus (Brink et al. 1995), burns (Reddi et al. 1985), liver or biliary disease (Brink et al. 1995), renal failure (Dan et al. 1992), or long-term catheterization (Bhandari et al. 2000).
There are several reports indicating infection due to S. putrefaciens following exposure to ma-rine life and the marine environment such as shellfish contact (Heller et al. 1990), bathing in the sea (Papanaoum et al. 1998), traumatic spike injury from a fish (Chen, 1997) and living on a boat (Yohe et al. 1997).
S. putrefaciens is being increasingly recog-nized as an opportunistic pathogen and as a cause of human infection (Brink et al, 1995; Chen et al, 1997; Leong et al, 2000). Detecting and identi-fying marine food spoilage organisms and op-portunistic pathogens in association with foods is therefore of importance.
Colonies of S. putrefaciens are characteristi-cally salmon colored. The ability of S. putrefa-ciens to form H2S was found to serve as a useful criterion of identity for the organism from marine fish with the use of pour plates of Peptone-Iron Agar (PIA; Levin, 1968). Subsurface colonies appear intensely black, whereas surface colonies usually have a black central area surrounded by a salmon colored peripheral area of growth. The ability of isolates to reduce trimethylamine oxide (TMA-O) to trimethylamine (TMA), to produce extracellular DNase and protease, to produce or-nithine decarboxylase, and to possess a mols% G+C content of 43 to 48% are additional criteria of identity (Levin, 1972; Levin, 1975; Gilardi, 1985; Vogel et al. 1997).
For over three decades S. putrefaciens has been recognized as a genetically heterogeneous species. Owen et al. (1978) showed that the spe-cies comprised at least four clearly separate ge-nomic groups, I to IV. One of the S. putrefaciens genomic groups (IV) has been reclassified as an additional species designated S. alga (Simidu et al (1990). Ziemke et al. (1998) designated Owen's genomic group II as a new species, S. baltica. Venkateswaran et al. (1999) designated Owen's group II as strains of a new species S.oneidensis. Owen's group I strains are highly related to ATCC 8071 and should be considered as the true S. purefaciens according to Tryfino-poulou et al. (2007). On the basis of phenotypic properties, isolates of S. putrefaciens can be eas-ily misidentified as S. alga (Vogel et al. 1997).
An inland fresh water tilapia farm located in Amherst, Massachusetts, U.S.A. was used as the source for all samples. This tilapia farm has cer-tain unique features including nitrifying tanks housing sand particles coated with nitrifying bacteria (Nitrosomonas europa and Nitrobacter winogradskyi) for conversion of ammonia se-quentially to nitrite and then to nitrate. The re-sulting nitrate is then consumed by hydroponi-cally grown basil plants. This allows the fish tank water to be constantly re-circulated with a loss of no more than about 2% of the total tank water daily.
In the present study isolates of Shewanella de-rived from various locations of this tilapia fish farm were characterized on the basis of a number of biochemical tests and the determination of the moles % G+C content of DNA. In addition, iso-lates were characterized with respect to RAPD analysis. The details regarding the operation of this tilapia plant and a flow diagram have pre-viously been described (Lu and Levin, 2007).
Materials and Methods
Sources of Shewanella isolates
All samples for bacterial analysis were ob-tained and processed in 2003. Approximately 75 ml samples of tilapia fish tank water derived from 180,000 gallons of tilapia farm water in 250,000 gallon tanks maintained at 30oC were collected in sterile 100 ml milk dilution bottles. All commercial culture media were Difco. Within 1 hr after collection the samples were de-cimally diluted in Tryptic Soy Broth (TSB) and 0.1 ml of each dilution surface smeared in tripli-cate onto Tryptic Soy Agar (TSA) plates for de-termination of total aerobic CFU. Plates were in-cubated at 32oC for 24 hr. Salmon pigmented colonies regarded tentatively as Shewanella sp. were picked and streaked onto TSA plates. Iso-lated colonies were then picked and transferred to TSA slants for incubation and refrigerated sto-rage.
Sand particles suspended in water from the nitrifying tank were collected in a 400 ml sterile beaker, covered with sterile aluminum foil, and then transported to the laboratory. The entire beaker of sand was poured onto sterile filter pa-per in a sterile Buchner funnel and then washed twice with 100 ml of sterile TSB to remove un-attached bacteria using a vacuum filtration sys-tem. Wet sand (15.0 g) was then blended with 100 ml of TSB for 1 min to detach bacterial cells from the sand particles. Decimal dilutions were then prepared and plated as above.
50 g of basil roots were blended with 250 ml of TSB for 30 sec. Decimal dilutions were pre-pared and then cultured for CFU as described above.
Physiological and biochemical Characteri-zation of the isolates
Characterization of all the Shewanella isolates was achieved as described below. Cell morphol-ogy, motility, gram stain, cytochrome oxidase, catalase, liquefaction of gelatin, proteolysis of casein, hydrolysis of DNA, reduction of nitrate, gas production from dextrose and were per-formed according to Burnett et al. (1957) and Pelczar and Reid (1958). Reduction of TMA-O to TMA was performed with 10 ml of TSB in tubes containing 0.01M TMA-O as previously de-scribed (Wood and Baird, 1943). TMA produc-tion after 2 days of incubation at 32oC was ex-amined by the method of Laycock and Reiger (1971).
Lecithinase production was determined by using nutrient agar supplemented with 10% (v/v) egg-yolk saline solution (50% v/v) (Stenstrom, 1990). Ornithine decarboxylase was detected by adding 2.0% DL-ornithine monohydrochloride to Decarboxylase Base Moeller (Difco) sealed with sterile mineral oil. Haemolytic activity was ob-served on plates of TSA supplemented with 5% defibrinated sheep blood. All strains were tested for the ability to grow at 4oC by incubating in-oculated tubes of TSB without dextrose for 12 days. Growth at 37oC was assessed by inoculat-ing PIA slants followed by incubation for 11 days. Growth at 42oC was assessed by inoculat-ing TSA plates followed by incubation for 1 day. H2S production was determined by stab inocu-lating PIA deeps followed by incubation at 32oC for 2 days. Growth on plates of Salmonella-Shi-gella (SS) Agar was determined after 7 days in-cubation at 37oC. Tolerance to NaCl was deter-mined in tubes holding 10 ml of TSB broth con-taining 6.0% and 10% NaCl incubated at 20oC for up to 12 days (Levin, 1972; Gilardi et al. 1991; Nozue et al, 1992; Vogel et al. 1997; Ven-kateswaran et al. 1999).
Acid production from glucose was assessed using the medium of Hugh and Leifson (1953) with aerobic and anaerobic (tubes sealed with 0.5 cm of sterile mineral oil) incubation after 3 days at 32oC.
Resistance to O/129 (2,4-diamino-6,7-diiso-propylpteridine, Sigma) was determined on in-oculated smear plates of TSA agar using the disk diffusion method. Fifty μL of filter sterilized O/129 (150 mg/50 μL) were applied to 14 mm diam. filter paper discs placed onto the center of the plate followed by incubation at 32oC for 24 hr.
RAPD analysis
Cells from a single Shewanella colony were subcultured overnight onto a plate of TSA con-taining 0.5% dextrose at 32oC. One purified co-lony was picked and inoculated into 5 mL of TSB broth and incubated overnight at 32oC with rotary agitation (200 rpm). A portion of the broth cul-ture (1.5 mL) was centrifuged at 12,000 rpm (16,000 g) for 5 min and the pellet washed twice with 1 ml of deionized water (dH2O). The pellet was then resuspended in 250 μL of sterile dH2O and adjusted to an absorbance of 1.6 at 600nm in 1 cm path length cuvets. This suspension was used directly for RAPD amplification or kept fro-zen at -20oC until used.
Three decamer primers LMPB4 5’-AAGGATCAGC, LMPB1 5’-GGAACTGCTA (Mazurier et al. 1992), and LMHLWL74 5’-ACGTATCTGC (Farber et al. 1994; Boerlin et al. 1995) synthesized by Sigma/Genesis were selected for RAPD typing of the Shewanella strains.
For the amplification procedure, 50 μL PCR reaction mixtures were prepared, each containing 1.25 U of Taq DNA polymerase (Qiagen, Fisher Scientific, cat. no. 201203), 2.5 mM MgCl2, 0.2 mM each dNTPs (dATP, dTTP, dCTP dGTP) (Takara Shuzo, Fisher Scientific, cat. no. TAK4030). 0.4 μM of the primer and 5 μL of cell suspension a (A600 = 1.6). The reaction mix-ture was cycled through the following tempera-ture profile: one cycle consisting of 94oC for 4 min, 35oC for 2 min and 72oC for 2 min followed by 43 cycles consisting of 94oC for 1 min, 35oC for 2 min and 72oC for 2 min. The final cycle was: 94oC for 1 min, 35oC for 2 min and 72oC for 10 min. All amplifications were performed in a DNA thermocycler (Techgene Ltd., Cambridge, UK).
For pattern analysis, 14 μL of the amplified DNA products were loaded and resolved on 1.6 % agarose gels by electrophoresis running at 5V/cm for 1.5 hr with 1 5 TBE running buffer (89 mM Tris-Base, 89 mM boric acid, 2 mM EDTA, pH8.4). The gels were stained with ethi-dium bromide (1 μg/mL) for 30 min. A DNA ladder ranging from 50 - bp to 2000 - bp (Sigma, cat. no. P9577) was included on each gel for molecular weight markers. Banding patterns were visualized with a UV transilluminator and photo-graphed with a digital camera with orange filter.
DNA extraction and mols% G+C determination
Cells were harvested from 1L flasks contain-ing 200 ml of TSB with 0.5% dextrose incubated overnight at 32oC with rotary agitation (200 rpm). DNA extraction was as previously de-scribed (Marmur, 1961; Levin, 1972).
The DNA melting point (Tm value) was de-termined as described by Marmur and Doty (1962) using an electrically heated cuvette and a calibrated thermistor temperature indicator and probe (Levin, 1969). The Mols% G+C content of DNA was calculated from the equation: %G+C = (Tm-53.9)/0.41 (Marmur and Doty, 1962). Escherichia coli K12 with a previously reported mols% G+C of 51.0% (Seidler et al. 1969) was used as a reference standard.
Results and Discussion
Incidence of Shewanella from Tilapia fish farm and characteristics of isolates
A total of 23 Shewanella strains were isolated from various sources of the tilapia fish farm on different dates (Table 1). The results in Table 1 indicate that strains of Shewanella were found to constitute 4.0%, 70%, and 9.9% of the bacterial flora in the water from the same tilapia tank at different sampling dates. A single sampling of the sand in the nitrifying tank yielded only one isolate of Shewanella (0.87%). The total aerobic bacterial counts in the water were found to be 2.0x104 CFU/mL, 5.8x104 CFU/mL, 8.8x104 CFU/mL for March 5, Sept. 9, and Oct. 8 respec-tively (Table 1). The CFU counts of sampled water from the same fish tank appeared to in-crease with each sampling, presumably due to a significant increase in the size of the fish and amount of fecal load with time. Shewanella was not found on the roots of the hydroponically grown basil. This observation suggests that there might be bacterial species on the roots that are either inhibitory or exclusionary to Shewanella. The observation that Shewanella from the fish tank water sample on Sept. 9 constituted 70% of the total aerobic bacterial population and thereby dominated the tilapia tank water may reflect an unknown nutritional dynamic.

Table 1: Shewanella from a tilapia fish farm.
The Shewanella isolates from March 5 were designated 26Ft, 46Ft, 65Ft and 93Ft, from Sept. 9 Ft1, Ft2, Ft3, Ft4, Ft5, Ft6, Ft7, and from Oct. 8 10ft, 11ft, 12ft, 21ft, 24ft, 42ft, 43ft, 44ft, 52ft, 69ft, 114ft. The single Shewanella isolate from sand in the nitrifying tank on July 9 was desig-nated 60sp (Table 2, Table 3).

Table 2: Characteristics of Shewanella isolates
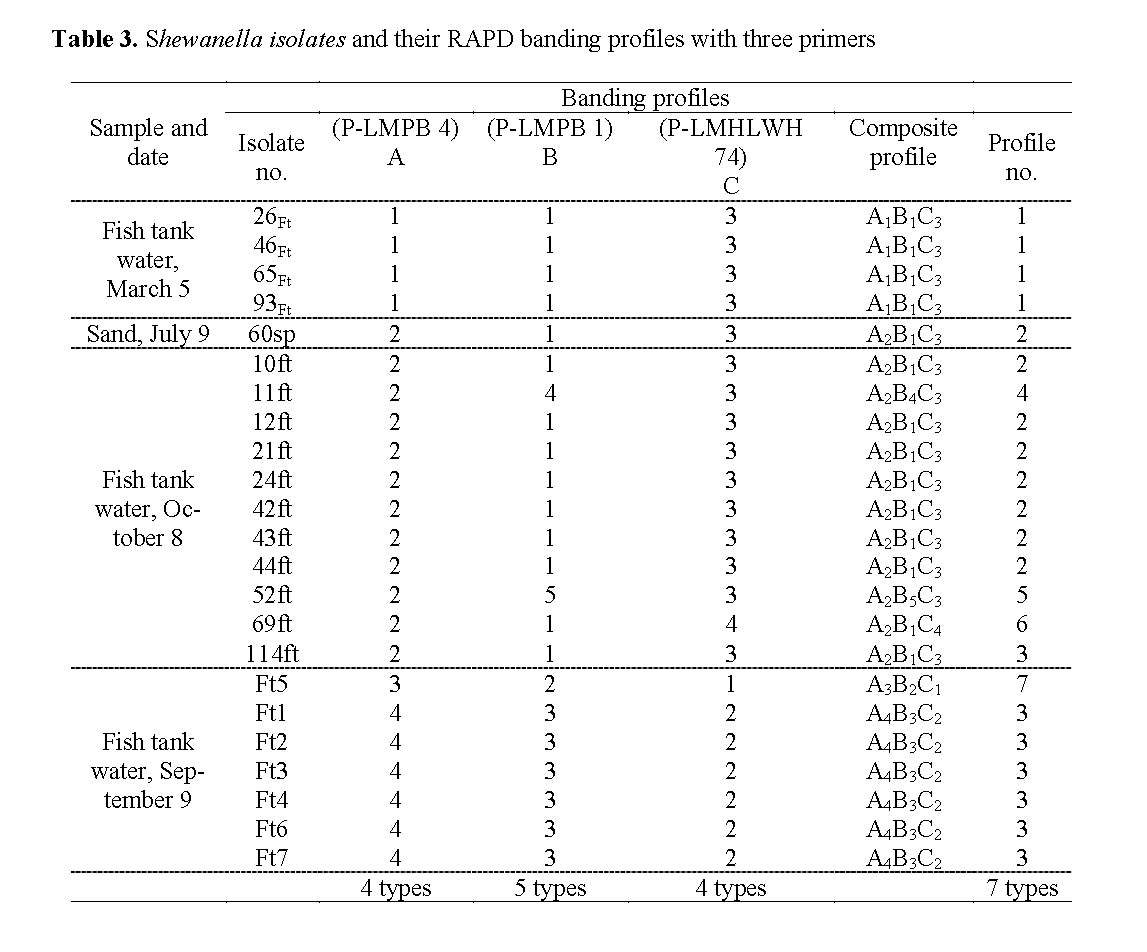
Table 3: Shewanella isolates and their RAPD banding profiles with three primers
All presumptive Shewanella strains isolated were Gram-negative, and composed of rod-shaped motile cells. Young colonies grown on TSA plates containing 0.5% dextrose were sal-mon colored.
Growth occurred at 37oC on PIA and at 42oC on TSA containing 0.5% dextrose, but not at 37oC on SS Agar with all 23 isolates (Table 2). All 23 Shewanella isolates produced hydrogen sulfide and grew at 4oC both in TSB broth with-out dextrose and on PIA slants (Table 2). Among the 23 isolates, the only ones able to grow in 6% NaCl were six of the seven from tilapia tank wa-ter obtained on Sept. 9. None of the 23 isolates were able to grow in 10% NaCl (Table 2).
All the strains of Shewanella were positive for production of catalase, cytochrome oxidase, DNase, proteinase (casein proteolysis), gelatinase (gelatin liquefaction), and ornithine decarbox-ylase. They were all negative for production of lecithinase. They were non-fermentative and no acid was oxidatively produced from dextrose in the medium of Hugh Leifson (Hugh et al, 1953) after 2 to 7 days of incubation but they did reduce TMA-O to TMA, and also reduced nitrate to ni-trite (Riley et al. 1972; Simidu et al. 1990; Reid et al. 1999; Venkateswaran et al. 1999). They were resistant to O/129 and exhibited β-haemoly-sis on sheep blood agar (Table 2). Although he-molysis was obscure on the first day of incuba-tion, it became obvious on the following days at 32oC.
RAPD banding profiles with three primers
Figure 1 presents the RAPD banding profiles for each of the seven isolates of Shewanella from fish tank water on Sept. 9 with the three random primers. Two RAPD profiles were collectively obtained. The isolate Ft5 was consistently distin-guished from the other six strains (which yielded identical profiles) with each of the 3 primers.
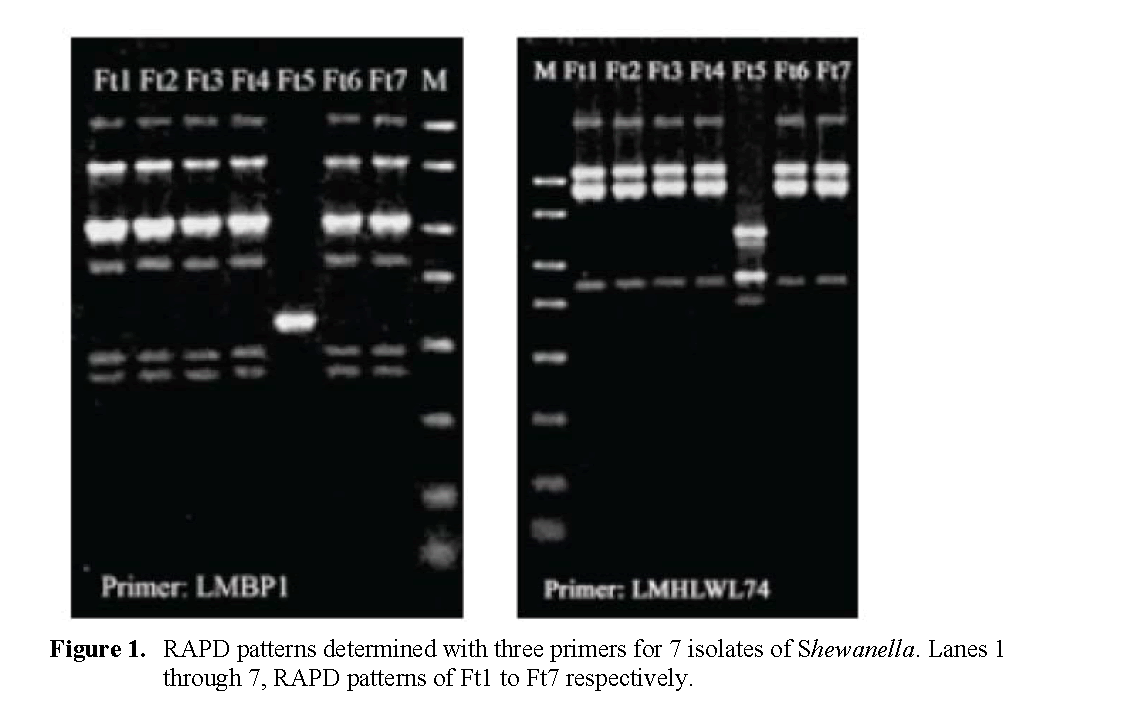
Figure 1: RAPD patterns determin ed with three primers for 7 isolates of Shewanella. Lanes 1 through 7, RAPD patterns of Ft1 to Ft7 respectively.
The additional 16 Shewanella strains were also subjected to RAPD analysis with the three primers. The RAPD profiles obtained were given arbitrary numerical designations shown in Table 3. From the 23 strains examined, 4, 5 and 4 dif-ferent RAPD profiles (Figure 2) were observed with primers LMPB1, LMPB4, LMHLWL74 respec-tively. There is a consensus band at about 800-bp with primer LMPB4 and at 900-bp with LMHLWL74 respectively. Based on RAPD analysis the 23 strains of Shewanella could be broken down into seven separate composite RAPD types with the use of the 3 primers (Table 3). This indicated that strains of Shewanella from the tilapia farm were genotypically heterogene-ous but with same common clones predomi-nantly.
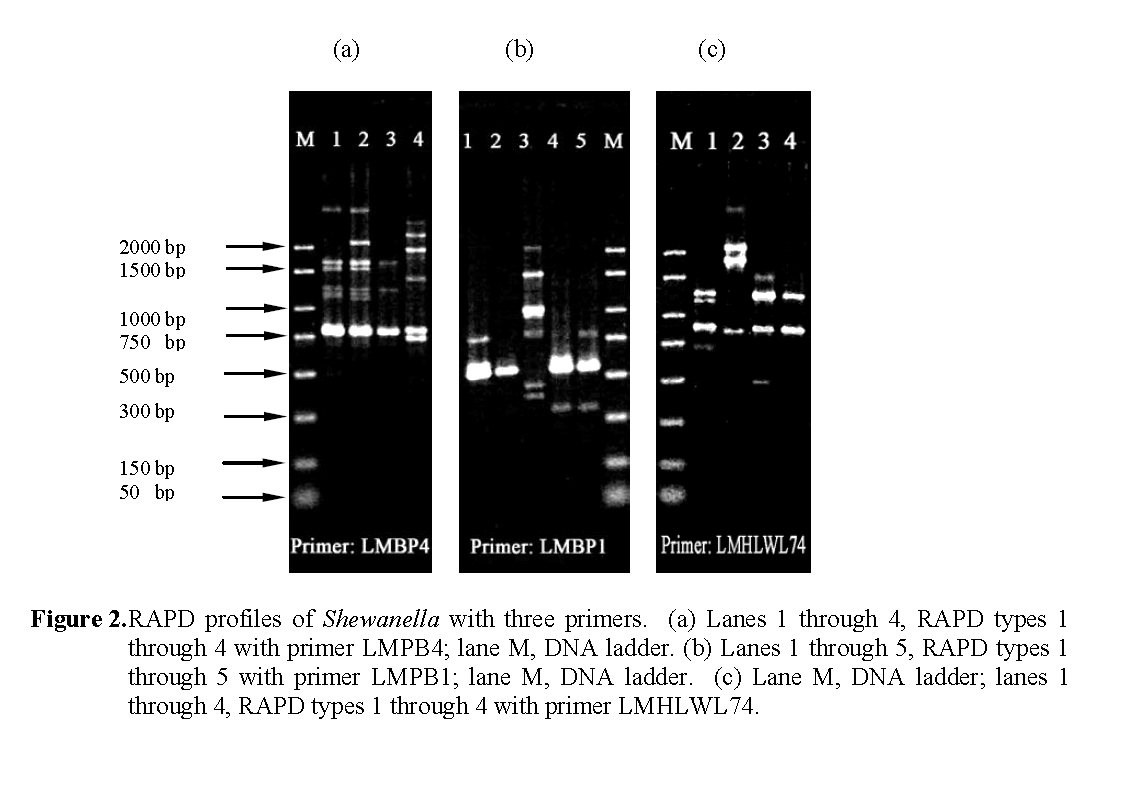
Figure 2: RAPD profiles of Shewanella with three primers. (a) Lanes 1 through 4, RAPD types 1 through 4 with primer LMPB4; lane M, DNA ladder. (b) Lanes 1 through 5, RAPD types 1 through 5 with primer LMPB1; lane M, DNA ladder. (c) Lane M, DNA ladder; lanes 1 through 4, RAPD types 1 through 4 with primer LMHLWL74.
The RAPD types of Shewanella were found to be different at different sampling dates except for the one isolate (60sp) from the sand particle of the nitrifying tank that corresponded to the domi-nant type 2 profiles of the fish tank water isolate of Oct. 8, 2003.
Determination of moles % G+C Content
The moles% G+C of the different RAPD types ranged from 45.5 ± 0.6 to 48.9 ± 0.6 re-spectively (Table 4). Levin (1972) previously dif-ferentiated intense fish spoilage isolates of P. putrefaciens and human clinical isolates into two groups: one group with a low moles % G+C content of 43.8 to 47.8% and the other group with a high moles % G+C content of 51.9 to 55.0%. He reported that the low G+C group was domi-nated by environmental and refrigerated food isolates, but did contain several human clinical isolates. Our results were similar to those he re-ported for fishery isolates and indicated that they were not S. alga isolates which have a high moles % G+C content of 52 to 54%, are able to grow on SS Agar, and in 10% NaCl but are unable to grow at 4 oC (Nozue et al, 1992; Vogel et al, 1997; Venkateswaran et al, 1999).
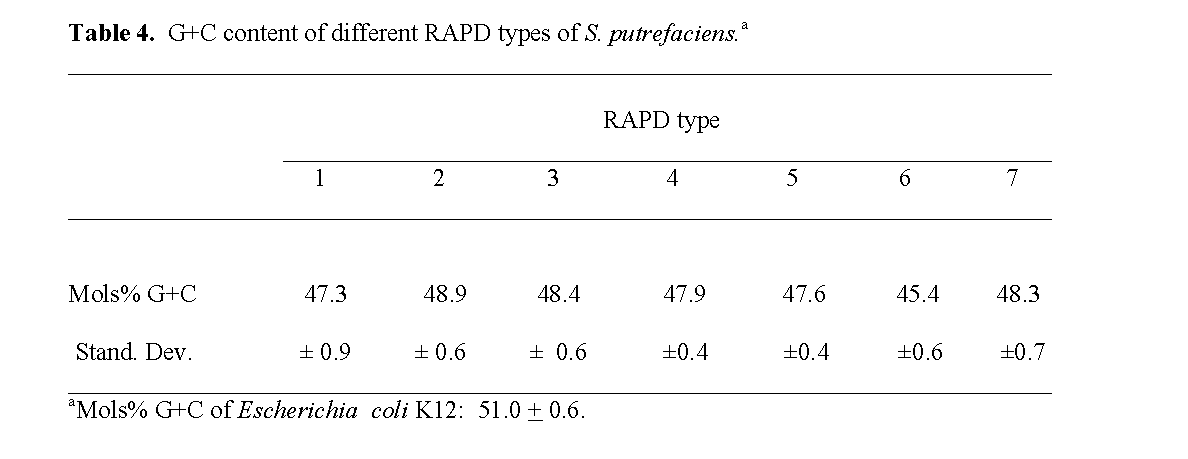
Table 4: G+C content of different RAPD types of S. putrefaciens.a
Sakata et al (1984) reported that the predomi-nant bacterial genera in the intestine of fresh wa-ter reared tilapia were Vibrio, Aeromonas and Pseudomonas. The present study is the first re-port that Shewanella can dominate the aerobic bacterial flora of a fresh water tilapia fish farm.
Basil roots failed to yield Shewanella. This might be due to some antagonistic activity. Pre-vious studies have reported that isolates of some Pseudomonas are capable of inhibiting the growth of S. putrefaciens (Gram, 1993; Gram and Melchiorsen, 1996).
Nozue et al (1992) attempted to clarify the physiological differences between S. alga and S. putrefaciens which are very similar. Thirty six strains of S. alga and 41 strains of S. putrefaciens were phenotypically and molecularly characte-rized. The two species can presumably be diffe-rentiated on the basis that S. alga grows at 42oC (Nozue et al, 1992; Khashe et al, 1998), and in 10% NaCl (Vogel et al, 1997; Venkateswaran et al, 1999) (Nozue et al, 1992; Khashe et al, 1998) while S. putrefaciens does not. In addition, strains of S. alga have a moles% G+C content of 52 to 54% in contrast to strains of S. putrefaciens that have a range of 45 to 48% (Nozue et al, 1992).
Tryfinopoulou et al. (2007) found that 16S rRNA sequence similarities (>97%) and pheno-typic similarities of S. baltica, S. purefaciens, and S. oneidensis made it difficult to allocate their Shewanella isolates from Aegean sea fish to a specific species .
In the present study the isolates of Shewanella from the tilapia farm had a G+C content ranging from 45.5 to 48.9%, were ?-hemolytic on sheep blood agar, grew at 4oC, 37oC, and 42oC, did not grow on SS agar, and did not produce lecithinase. Six of the seven Shewanella isolates from tilapia tank water obtained on Sept. 9. were able to grow in 6% NaCl but did not grow in 10% NaCl. An examination of the data in Table 5 makes it diffi-cult to allocate these isolates to any one specific species of Shewanella.

Table 5: Physiological characteristics of groups I, II, III, and IV derived from the grouping of S. pu-trefaciens isolates by Owen et al. (1978) and tilapia farm isolates of this study.a
Conclusions
The percent of Shewanella in tilapia farm tank water was found to vary from 4.0% to 70% de-rived from total aerobic plate counts. Hydro-ponically grown basil plants associated with the fish farm tank water failed to yield Shewanella isolates. This is the first report of a single clone of Shewanella numerically dominating the aero-bic bacterial flora of the tank water of a tilapia fish farm.
Acknowledgements
This research was supported by a Special Sea-food Safety Grant from the USDA.
1037
References
- Bhandari, S., Pan, T.L.T., Horvath, J., Tiller, D. (2000). CAPD, swimming in Shewanella, Nephrol Dial Transplant, 15: 1484-1485. doi:10.1093/ndt/15.9.1484
- nBoerlin, P., Bannerman, E., Ischer, F., Rocourt J., Bille, J. (1995). Typing Listeria monocyto-genes: a comparison of random amplifica-tion of polymorphic DNA with 5 other me-thods, Research in Microbiology, 146: 35-49. doi:10.1016/0923-2508(96)80269-5
- nBorch E., Kant-Muermans M.L., Blixt Y. (1996). Bacterial spoilage of meat and cured meat products, International Journal of Food Mi-crobiology, 33: 103-120. doi:10.1016/0168-1605(96)01135-X
- nBrink, A.J., van Staten, A., van Rensburg, A.J. (1995). Shewanella (Pseudomonas) putrefa-ciens bacteremia, Clinical Infectious Dis-eases, 20: 1327-1332
- nBurnett, G.W., Pelczar M.J., Conn, H.J. (1957). Preparation of media. In M.J Pelczar,. Jr., R.C. Bard, G.W. Burnett, H.J. Conn, R.D. DeMoss, E.E. Evans, M.W. Jennison, A.P. McKee, A.J. Riker, J.Warren, O.B. Weeks, F.A Weiss, eds. Manual of Microbiological Methods. pp. 37-63. McGraw-Hill, Inc., New York, Toronto, London:
- nCastell, C.H., Richards, J.F., Wilmot, I. (1949). Pseudomonas putrefaciens from cod fillets, Journal of the Fisheries Research Board of Canada, 7: 430-431
- nChai, T., Chen, C., Rosen, A., Levin, R.E. (1968). Detection and Incidence of Specific Species of Spoilage Bacteria on Fish, Ap-plied Microbiology, 16: 1738-1741
- nChen, Y.S., Liu, Y.C., Yen, M.Y., Wang, J.H., Wang, J.H., Wann S.R., Cheng, D.L. (1997). Skin and soft-tissue manifestations of She-wanella putrefaciens infection, Clinical In-fectious Diseases, 25: 225-229
- nDan, M., Gutman R., Biro, A. (1992). Peritonitis caused by Pseudomonas putrefaciens in pa-tients undergoing continuous ambulatory pe-ritoneal dialysis. Clinical Infectious Dis-eases, 14: 359-360
- nDerby, H.A., Hammer, B.W. (1931). Bacteriol-ogy of butter. IV. Bacteriological studies on surface taint butter. Iowa Agric. Exp. Stat. Res. Bull. 145: 387-416
- nFarber, J.M., Addison, C.J. (1994). RAPD typing for distinguishing species and strains in the genus Listeria, Journal of Applied Bacteri-ology, 77: 242-250
- nGilardi, G.L. In Lennette, E.H., Balows, A., Hausler, W.J., Jr., and Shadomy, H.J. (1985). Pseudomonas. Manual of Clinical Microbiology, 4th ed. pp. 350-372. Wash-ington, DC: American Society for Microbi-ology
- nGilardi, G.L., In: Balows, A., Hausler, W.J. Jr, Herrmann, K.L., Isenberg, H.D., Shadomy, H.J. (1991). Pseudomonas and related ge-nera. Manual of Clinical Microbiology. 5th ed. pp. 429-441. Washington, DC: American Society for Microbiology
- nGram, L. (1992). Evaluation of the bacteriologi-cal quality of seafood, International Journal of Food Microbiology, 16: 25-39. doi:10.1016/0168-1605(92)90123-K
- nGram, L., Melchiorsen, J. (1996). Interaction between fish spoilage bacteria Pseudomonas sp. and Shewanella putrefaciens in fish ex-tracts and on fish tissue, Journal of Applied Bacteriology, 80: 589-595
- nGram, L. (1993). Inhibitory effect against pa-thogenic and spoilage bacteria of Pseudo-monas strains isolated from spoiled and fresh fish, Applied and Environmental Mi-crobiology, 59: 2197-2203
- nGram, L, Huss, H.H. (1996). Microbiological spoilage of fish and fish products, Interna-tional Journal of Food Microbiology, 33: 121-137. doi:10.1016/0168-1605(96)01134-8
- nHeller, H., Tortora, G., Burger, H. (1990). Pseudomonas putrefaciens bacteremia asso-ciated with shellfish contact, The American Journal of Medicine, 88: 85-86
- nHolmes, B., Lapage, S.P., Malnick, H. (1975). Strains of Pseudomonas putrefaciens from clinical material, Journal of Clinical Pa-thology, 28: 149-155
- nHugh, R., Leifson, E. (1953). The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various Gram negative bacteria, The Journal of Bacteriology, 66: 24-26
- nJørgensen, B.R., Huss, H.H. (1989). Growth and activity of Shewanella putrefaciens isolated from spoiling fish, International Journal of Food Microbiology, 9: 51-62
- nKhashe, S., Janda, J.M., (1998). Biochemical and Pathogenic Properties of Shewanella alga and Shewanella putrefaciens, Journal of Clinical Microbiology, 36: 783-787
- nLaycock, R.A., Reiger, L.W. (1971). Trimethy-lamine-producing bacteria on haddock (Melanogrmmus aeglefinus) fillets during refrigerated storage, Journal of the Fisheries Research Board of Canada, 28: 305-309
- nLee, J.V., Gibson, D.M., Shewan, J.M. (1977). A numerical taxonomic study of Pseudomo-nas-like marine bacteria, Journal of Gen-eral Microbiology, 98: 439-451
- nLeong J., Mirkazemi M., Kimble F. (2000). Shewanella putrefaciens hand infection, Australian and New Zealand Journal of Surgery, 70: 816-817. doi:10.1046/j.1440-1622.2000.01962.x
- nLevin, R.E. (1968). Detection and incidence of specific species of spoilage bacteria on fish I. Methodology, Applied Microbiology, 16: 1734-1737
- nLevin R. E. (1969). Electrically Heated Cuvette Chamber for deoxyribonucleic acid melting point determinations. Applied Microbiology, 18: 528-530
- nLevin, R.E. (1975). Characteristics of weak-H2S-producing isolates of Pseudomonas putrefa-ciens from human infections Ant. van Leeu-wen, J. Microbiol. Serol. 41: 569-574
- nLevin, R.E. (1972) Correlation of DNA base composition and metabolism of Pseudomo-nas putrefaciens isolates from food, human clinical specimens, and other sources. Ant. van Leeuwen, J. Microbiol. Serol. 38: 121-127
- nLong, H.F., Hammer B.W. (1941). Classifica-tion of organisms important in dairy prod-ucts. III. Pseudomonas putrefaciens, Iowa Agric. Exp. Stat. Res. Bull, 285: 176-195
- nLu, S., Levin, R. (2008). Dominant bacterial Genera of a tilapia fish farm and RAPD typing of Vibrio isolates. Journal of Fishe-resSciences.com, 2: 183-195. doi:10.3153/jfscom.2008020
- nMacDonell, M.T., Colwell, R.R. (1985). Phylo-geny of the Vibrionaceae and recommenda-tion for two new genera, Listonella and Shewanella, Systematic and Applied Micro-biology, 6: 171-182
- nMarmur, J. (1961). A procedure for the isolation of deoxyribonucleic acid from micro-organ-isms, Journal of Molecular Biology, 3: 208-218
- nMarmur, J., Doty, P. (1962). Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature, Journal of Molecular Biology ,5: 109-118
- nMazurier, S., Wernars, K. (1992). Typing of Listeria strains by random amplification of polymorphic DNA (RAPD) analysis, Jour-nal of Medical Microbiology, 38: 322-327
- nMazurier, S.-I., Wernars, K. (1992). Typing of Listeria strains by random amplification of polymorphic DNA, Research in Microbiol-ogy, 143: 499-505. doi:10.1016/0923-2508(92)90096-7
- nNozue, H., Hayashi, T., Hashimoto, Y., Ezaki, T., Hamasaki, K. Ohwada, K. and Terawaki, Y. (1992). Isolation and characterization of Shewanella alga from human clinical spe-cimens and emendation of the description of S. alga Simidu et al. 1990. 335: Interna-tional Journal of Systematic Bacteriology, 42: 628-634
- nPagani, L., Lang, A., Vedovelli, C., Moling, O., Rimenti, G., Pristera, R. and Mian, P. (2003). Soft tissue infection and bacteremia caused by Shewanella putrefaciens, Journal of Clinical Microbiology,41: 2240-2241. doi:10.1128/JCM.41.5.2240-2241.2003
- nPapanaoum, K., Marshmann, G., Gordon, L.A., Lumb, R., Gordon, D.L. (1998). Concur-rent infection due to Shewanella putrefa-ciens and Mycobacterium marinum acquired at the beach, Australian Journal of Derma-tology, 24: 798-800
- nPelczar, M.J., Jr, Reid, R.D. (1958). Laboratory Exercises in Microbiology. pp. 71-74. McGraw-Hill, Inc. New York, Toronto, London
- nReddi, G.S., Shukl, N.P., Singh, K.V. (1985). Pseudomonas putrefaciens as a cause of in-fection in burn, Indian Journal of Pathology & Microbiology, 28: 303-308
- nRiley, P.S., Tatum, H.W., Weaver, R.E. (1972). Pseudomonas putrefaciens isolates from clinical specimens, Applied Microbiology, 24: 798-800
- nSeidler, R.J., Starr, M.P., Mandel, M. (1969). Deoxyribonucleic acid characterization of BdelloVibrios, Journal of Bacteriology, 100: 786-790
- nStenstrom, I.-M., Molin, G. (1990). Classifica-tion of the spoilage flora of fish, with special reference to Shewanella putrefaciens, Jour-nal of Applied Bacteriology, 68: 601-618
- nTryfinopoulou, P., Tsakalidou, E., Vancanneyt, M., Hoste, B., Swings, J., Nychas, G. (2007). Diversity of Shewanella population in fish Sparus aurata harvested in the Ae-gean Sea, Journal of Applied Bacteriology, 103: 711-721. doi:10.1111/j.1365-2672.2007.03355.x
- nVenkateswaran, K., Moser, D.P., Dollhopf, M.E., Lies, D.P., Saffarini, D.A., MacGregor, B.J., Ringelberg, D.B., White, D.C., Nishijima, M., Sano, H., Burghardt, J., Stackebrandt, E., Nealson. K.H. (1999). Polyphasic tax-onomy of the genus Shewanella and de-scription of Shewanella oneidensis sp. Nov, International Journal of Systematic Bacteri-ology, 49: 705-724
- nVon Graevenitz, A., Simon, G. (1970). Poten-tially pathogenic, nonfermentative, H2S-pro-ducing gram-negative rod (1b), Applied Microbiology, 19: 176
- nVogel, B.F., Jørgensen, K., Christensen, H., Ol-sen, J.E., Gram, L. (1997). Differentiation of Shewanella putrefaciens and Shewanella alga on the basis of whole-cell protein pro-files, ribotyping, phenotypic characteriza-tion, and 16S rRNA gene sequence analysis, Applied and Environmental Microbiology, 63: 2189-2199
- nWood, A.J., Baird, E.A. (1943). Reduction of trimethylamine oxide by bacteria. 1. The Enterobacteriaceae, Journal of the Fisheries Research Board of Canada, 6: 194-201
- nYohe, S., Fishbain, J.T., Andrew, M. (1997). Shewanella putrefaciens abscess of the lower extremity. Journal of Clinical Micro-biology, 35: 33-36.